We present SubGNN, a general method for subgraph representation learning. It addresses a fundamental gap in current graph neural network (GNN) methods that are not yet optimized for subgraph-level predictions.
Our method implements in a neural message passing scheme three distinct channels to each capture a key property of subgraphs: neighborhood, structure, and position. We have generated four synthetic datasets highlighting a specific subgraph property. By performing an ablation study over the channels, we demonstrate that the performance of individual channels aligns closely with their inductive biases.
SubGNN outperforms baseline methods across both synthetic and real-world datasets. Along with SubGNN, we have released eight new datasets representing a diverse collection of tasks to pave the way for innovating subgraph neural networks in the future.
Publication
Subgraph Neural Networks
Emily Alsentzer, Samuel G. Finlayson, Michelle M. Li, Marinka Zitnik
NeurIPS 2020 [arXiv] [poster]
@inproceedings{alsentzer2020subgraph,
title={Subgraph Neural Networks},
author={Alsentzer, Emily and Finlayson, Samuel G and Li, Michelle M and Zitnik, Marinka},
booktitle={Proceedings of Neural Information Processing Systems, NeurIPS},
year={2020}
}
Motivation
Encoding subgraphs for GNNs is not well-studied or commonly used. Rather, current GNN methods are optimized for node-, edge-, and graph-level predictions, but not yet at the subgraph-level.
Representation learning for subgraphs presents unique challenges.
- Subgraphs require that we make joint predictions over structures of varying sizes. They do not necessarily cluster together, and can even be composed of multiple disparate components that are far apart from each other in the graph.
- Subgraphs contain rich higher-order connectivity patterns, both internally and externally with the rest of the graph. The challenge is to inject information about border and external subgraph structure into the GNN’s neural message passing.
- Subgraphs can be localized or distributed throughout the graph. We must effectively learn about subgraph positions within the underlying graph.
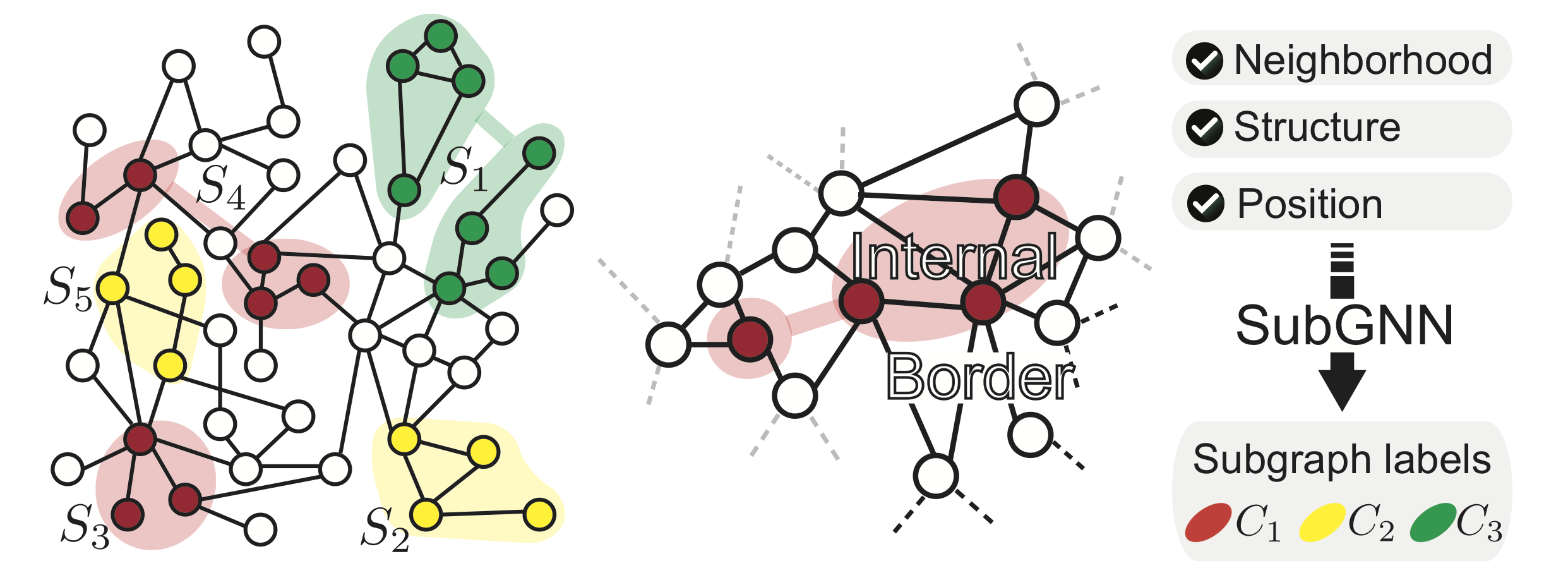
The following figure depicts a simple base graph and five subgraphs, each with different structures. For instance, subgraphs S2, S3, and S5 comprise of single connected components in the graph, whereas subgraphs S1 and S4 each form two isolated components. Colors indicate subgraph labels. The right panel illustrates an example of internal connectivity versus border structure
SubGNN framework
SubGNN takes as input a base graph and subgraph information to train embeddings for each subgraph. Each channel’s output embeddings are then concatenated to generate one final subgraph embedding.
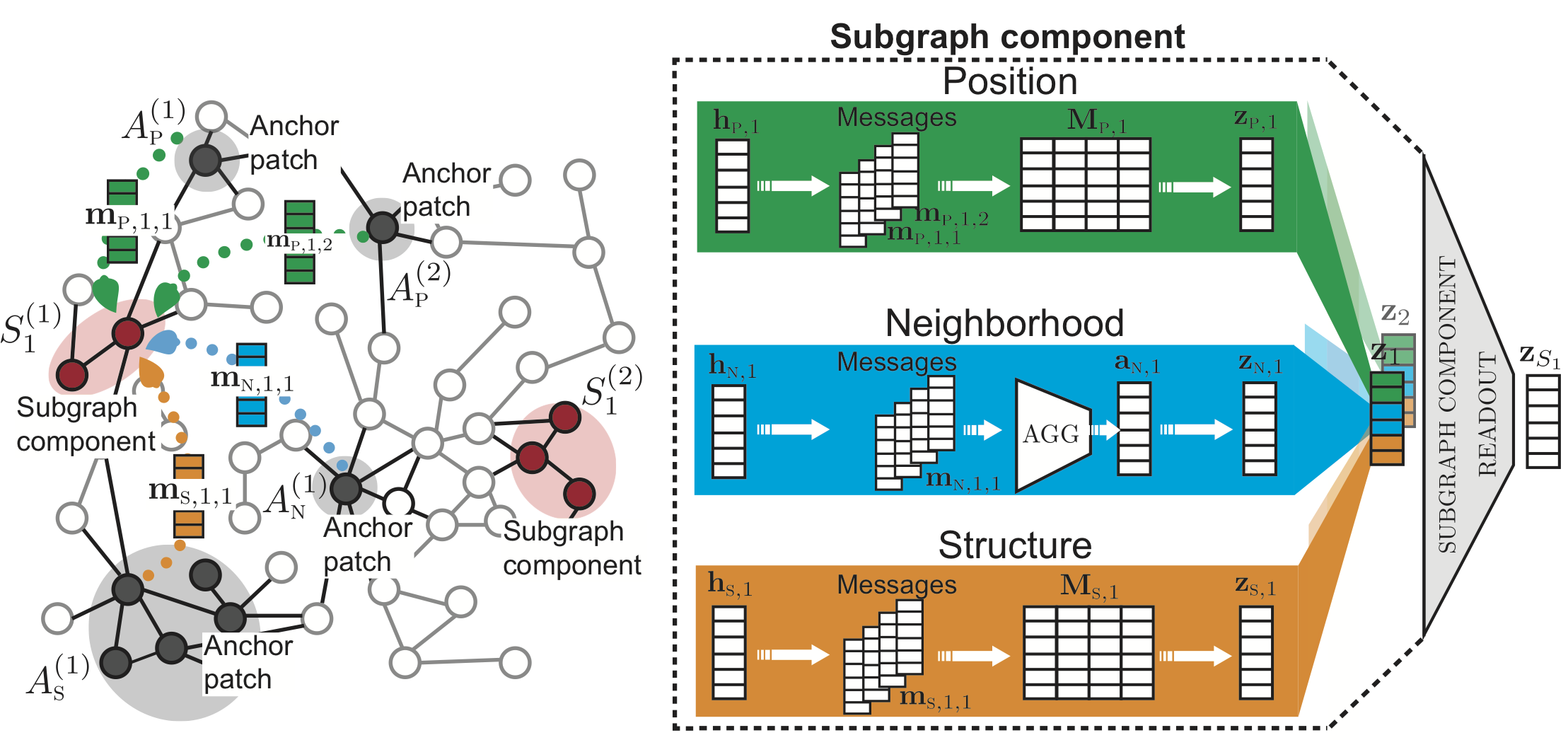
The figure above depicts SubGNN’s architecture. The left panel illustrates how property-specific messages are propagated from anchor node patches to subgraph components. The right panel shows the three channels designed to capture a distinct subgraph property.
Datasets
We present four new synthetic datasets and four novel real-world social and biological datasets to stimulate subgraph representation learning research.
Synthetic datasets
Each synthetic dataset challenges the ability of our methods to capture:
- DENSITY: Internal structure of subgraph topology.
- CUT RATIO: Border structure of subgraph topology.
- CORENESS: Border structure and position of subgraph topology.
- COMPONENT: Internal and external position of subgraph topology.
Real-world datasets
- PPI-BP is a molecular biology dataset, where the subgraphs are a group of genes and their labels are the genes’ collective cellular function. The base graph is a human protein-protein interaction network.
- UDN-METAB is a clinical dataset, where the subgraphs are a collection of phenotypes associated with a rare monogenic disease and their labels are the subcategory of the metabolic disorder most consistent with those phenotypes. The base graph is a knowledge graph containing phenotype and genotype information about rare diseases.
- UDN-NEURO is similar to UDN-METAB but for one or more neurological disorders (multilabel classification), and shares the same base graph.
- EM is a social dataset, where the subgraphs are the workout history of a user and their labels are the gender of the user. The base graph is a social fitness network from Endomondo.
Code
Source code is available in the GitHub repository.
Authors
- Emily Alsentzer (co-first authors)
- Samuel G Finlayson (co-first authors)
- Michelle M Li
- Marinka Zitnik