CLEF is a model for counterfactual reasoning, enabling precise, controlled edits to sequential data. Traditional sequence models can generate counterfactuals—answering "what if" questions—but they lack control over when and where changes happen. Existing methods either treat sequences as a whole or assume every variable is affected, making them unsuitable for real-world applications where interventions impact only specific elements at precise moments.
CLEF introduces a new approach by learning temporal concepts that determine how and when an intervention influences a sequence. This allows CLEF to make targeted edits while preserving unaffected data. We validate CLEF on cellular and patient trajectory datasets, showing that it accurately models gene regulation and medical interventions, improving immediate sequence editing accuracy by up to 36.01% over baselines.
Unlike previous methods, CLEF generates counterfactual sequences in a single step at any future time point, outperforming baselines by up to 65.71% in MAE. In a case study on type 1 diabetes, CLEF identified clinical interventions that could shift patient trajectories toward healthier outcomes, demonstrating its potential for real-world impact.
Motivation
Counterfactual reasoning is essential in biology and medicine, enabling us to explore “what if” scenarios that shape our understanding of cells, diseases, and treatments. Decisions in these domains must consider not only the choice of an intervention, such as a drug or a surgical procedure, but also its timing and frequency. Generating realistic counterfactual sequences requires precise and context-aware edits that respect temporal and structural constraints. Existing generative models in language and vision allow fine-tuned modifications through prompts, but they lack the ability to perform controlled edits in time-sensitive sequences.
Current approaches to controllable sequence editing struggle with two key limitations. First, they excel at immediate editing (modifying the next step in a sequence under a counterfactual condition) but fail to generate long-term counterfactual trajectories while maintaining causal consistency. Second, existing methods for counterfactual time series generation assume that interventions affect the entire sequence rather than allowing targeted edits to specific variables while preserving unaffected data. These limitations hinder applications in cellular reprogramming and patient trajectory modeling, where interventions must be precisely controlled in both timing and scope.
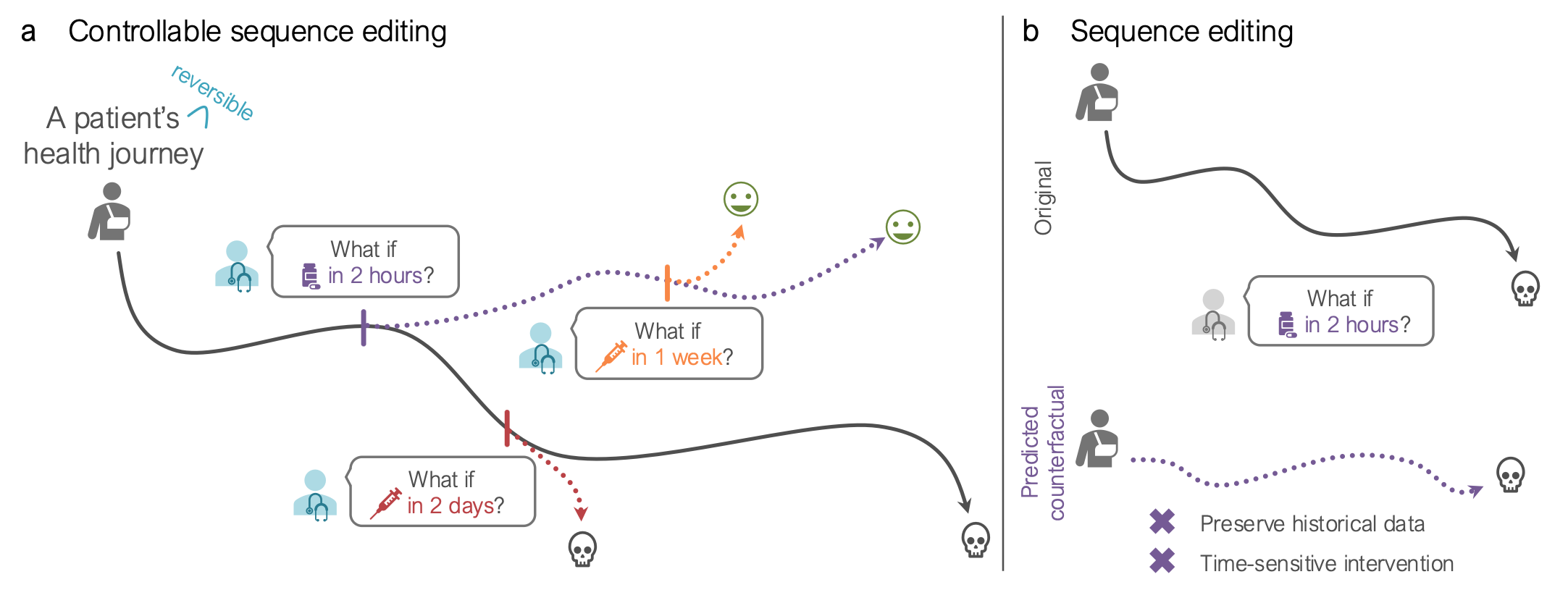
CLEF (Controllable Sequence Editing for Counterfactual Generation) is a model designed for precise counterfactual reasoning in biological and medical applications. Unlike traditional sequence models that modify an entire sequence globally, CLEF allows localized modifications at specific time steps, ensuring interventions only affect relevant portions of a sequence while maintaining causal consistency.
By leveraging these components, CLEF enables immediate and delayed sequence editing—allowing it to answer “what if” questions such as:
- What if a patient receives a treatment today versus next year?
- What if a genetic modification is introduced at a specific stage of cell development?
Overview of CLEF
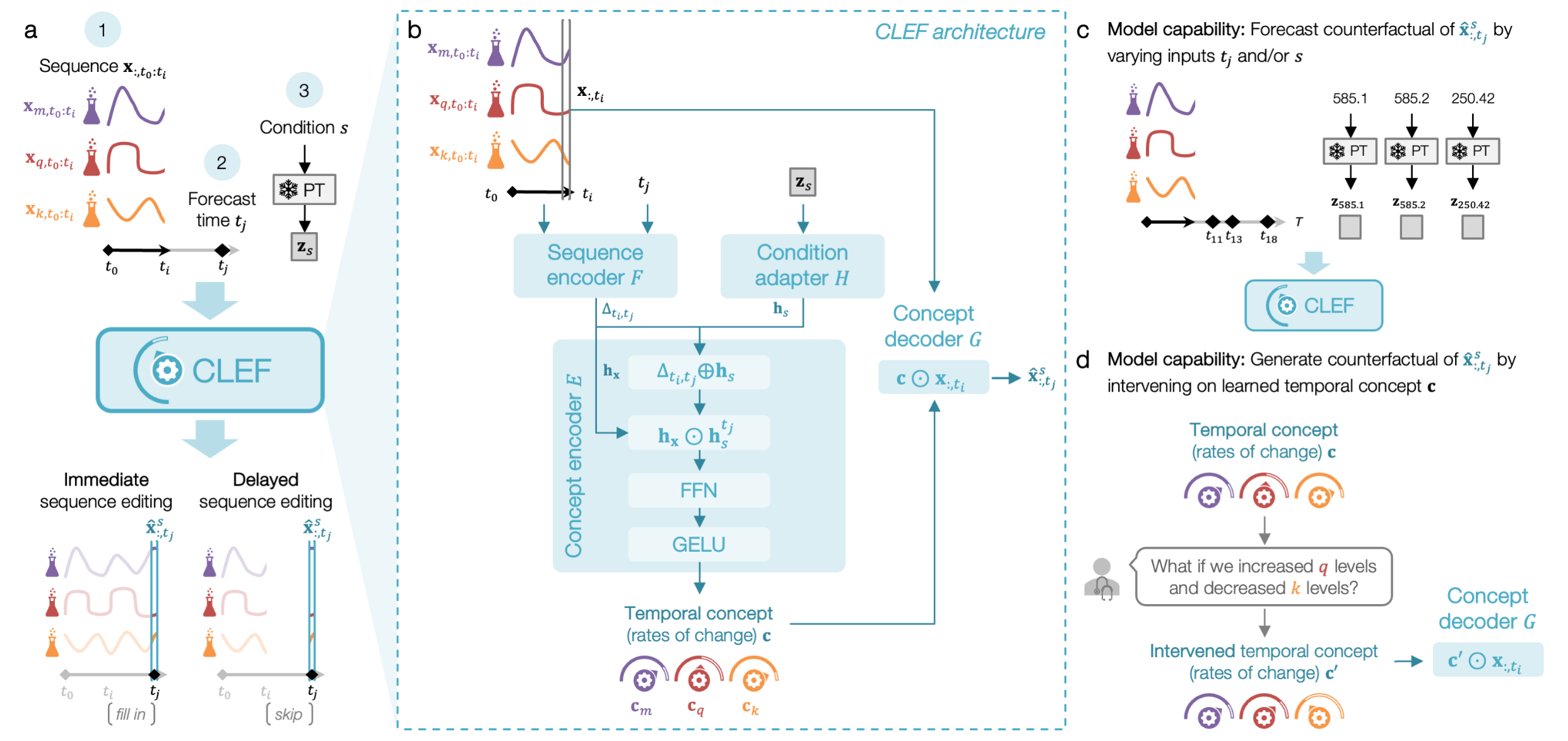
CLEF is designed to modify sequential data in response to user-specified conditions while ensuring causal consistency. Given an input sequence, a future time point, and a counterfactual condition, CLEF selectively edits relevant time steps while preserving the unaffected portions of the sequence. This enables reasoning about both immediate and delayed effects of interventions.
CLEF operates through four key components: a sequence encoder that extracts temporal patterns, a condition adapter that maps counterfactual inputs into latent representations, a concept encoder that learns temporal concepts, and a concept decoder that applies these learned patterns to generate counterfactual sequences. CLEF supports precise intervention at any time step, allowing for both short-term modifications and long-term outcome forecasting.
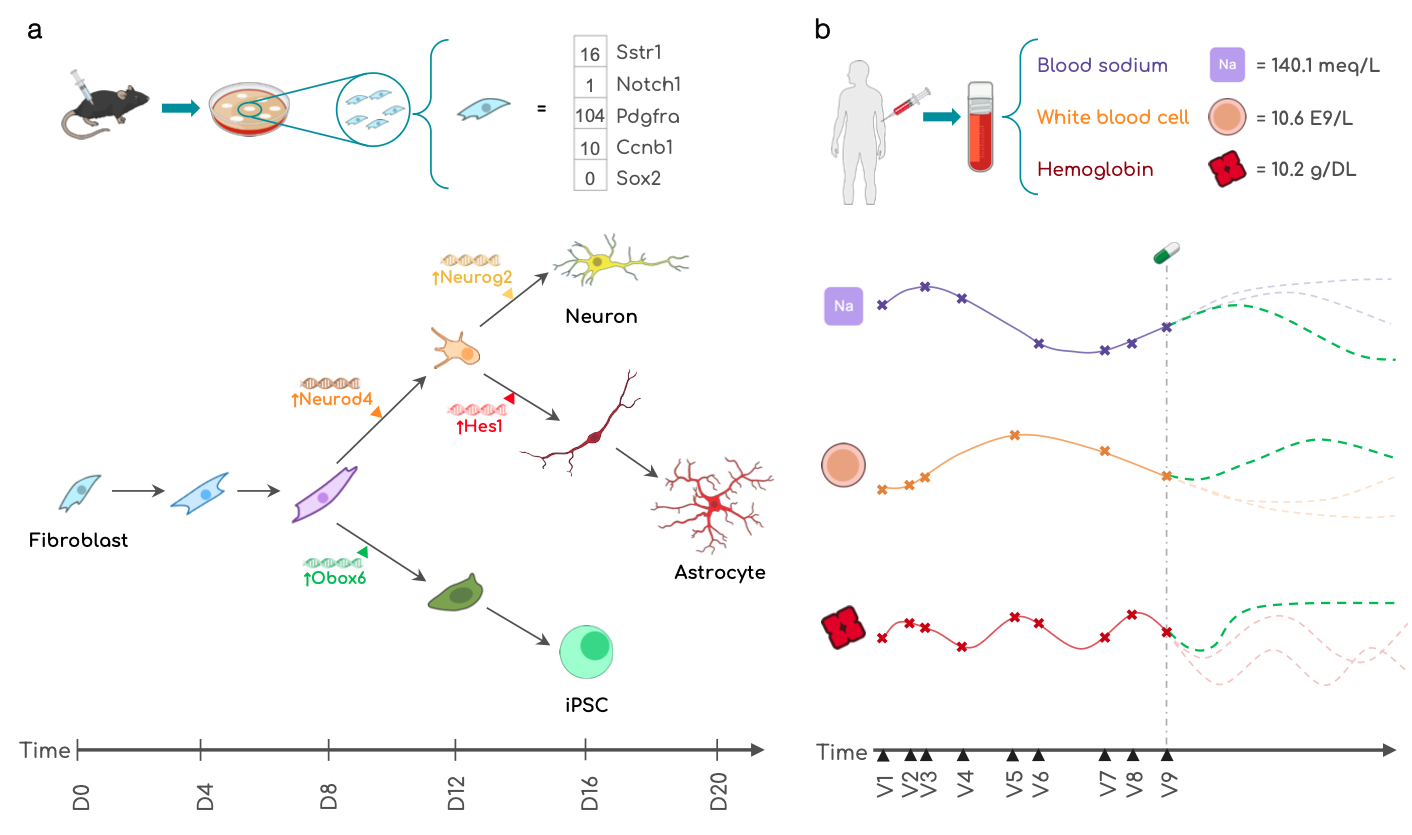
Experiments in cellular reprogramming and patient trajectory modeling
CLEF is evaluated on two critical applications: cellular reprogramming and patient health trajectories.
- In cellular reprogramming, CLEF models the effects of genetic perturbations, predicting how a cell’s developmental path changes when a transcription factor is activated at a specific time.
- In patient trajectory modeling, CLEF forecasts how clinical interventions—such as medication administration—affect future lab test results while preserving historical medical data.
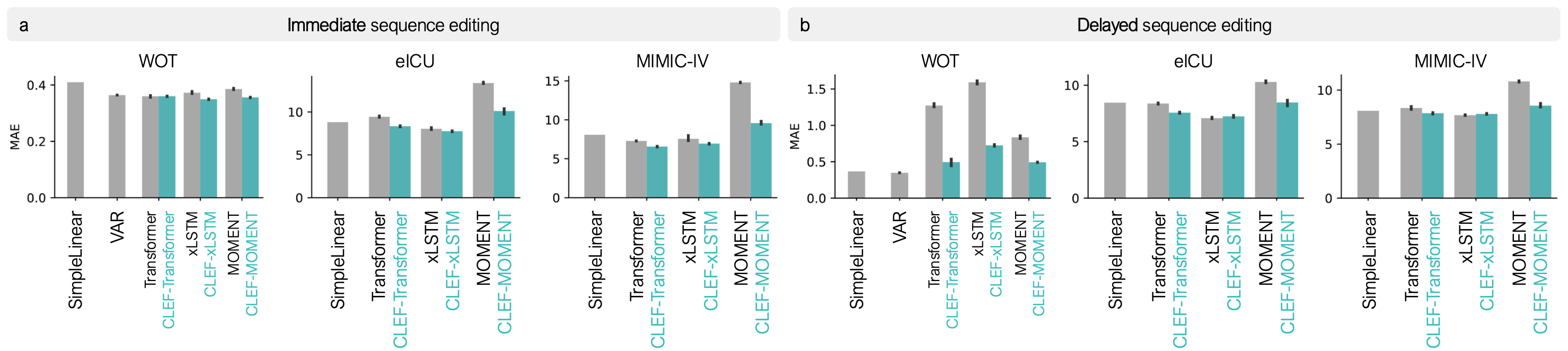
CLEF outperforms existing models in both tasks. It provides more accurate counterfactual predictions amd performs one-step forecasting of future states, reducing errors that accumulate over time in autoregressive models. In real-world patient datasets, CLEF generalizes well to unseen cases, demonstrating its robustness in clinical applications.
CLEF achieves state-of-the-art performance in counterfactual sequence editing. In immediate sequence editing, CLEF improves mean absolute error (MAE) by up to 36.01% over baselines. In delayed sequence editing, where counterfactual trajectories must be generated for future time steps, CLEF outperforms existing methods by up to 65.71% in MAE. These improvements are consistent across multiple datasets, including cellular reprogramming and patient lab test records, highlighting CLEF’s effectiveness in both short-term and long-term counterfactual reasoning.
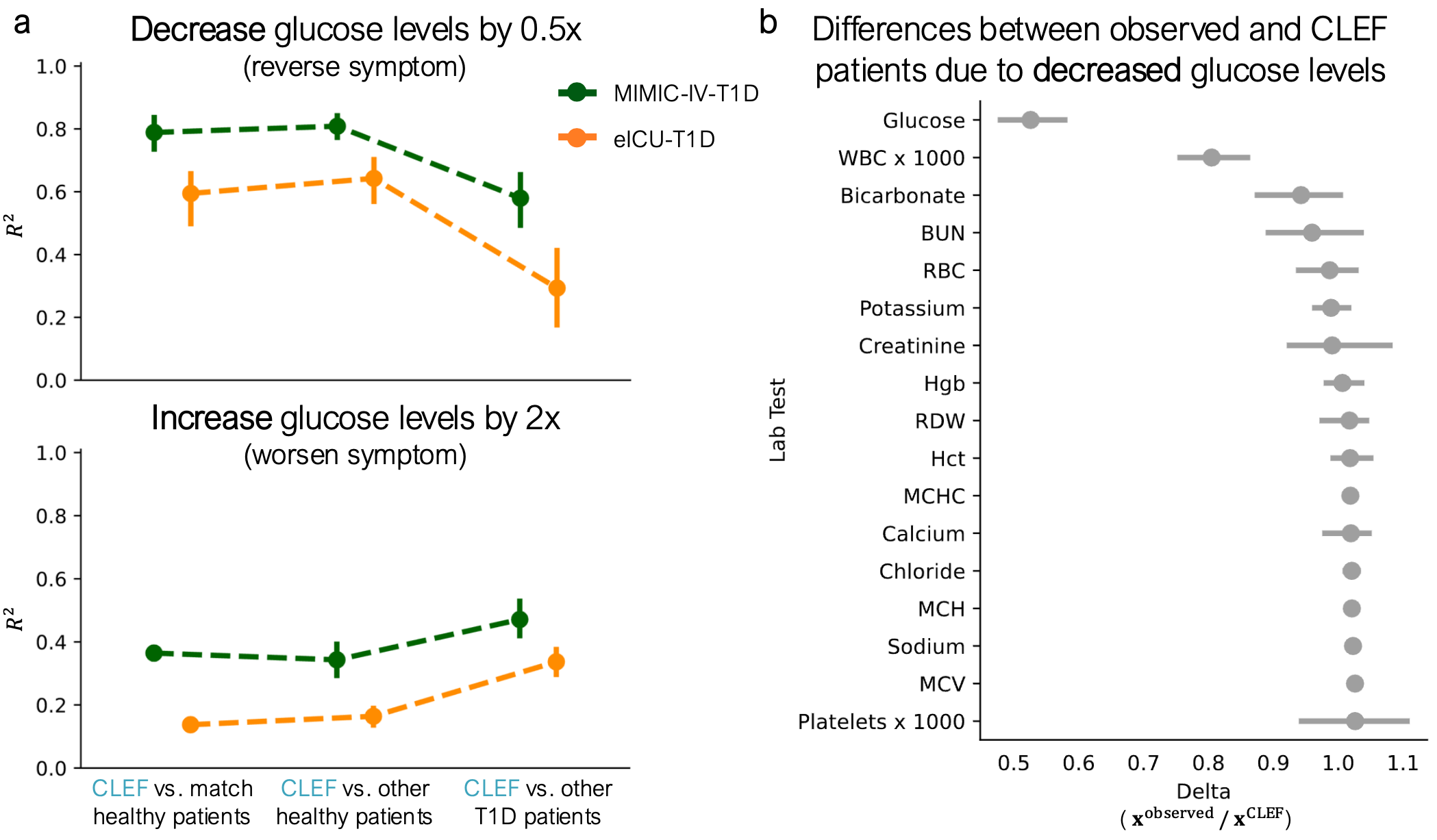
Counterfactual generation of glucose levels in patients with diabetes
CLEF enables precise intervention on learned temporal concepts, allowing for clinically meaningful counterfactual simulations. In a study on type 1 diabetes patients, CLEF was used to modify glucose levels, generating counterfactual trajectories that aligned more closely with healthy patient outcomes. Lowering glucose levels in the model led to a corresponding reduction in white blood cell counts, reflecting real physiological relationships.
Similarly, increasing glucose levels produced trajectories that more closely resembled those of other diabetic patients. These results demonstrate CLEF’s ability to generate interpretable counterfactuals that capture the effects of medical interventions, offering a powerful tool for exploring treatment strategies.
Publication
Controllable Sequence Editing for Counterfactual Generation
Michelle M. Li, Kevin Li, Yasha Ektefaie, Shvat Messica, Marinka Zitnik
In Review 2025 [arXiv]
@article{li2025controllable,
title={Controllable Sequence Editing for Counterfactual Generation},
author={Li, Michelle M and Li, Kevin and Ektefaie, Yasha and Messica, Shvat and Zitnik, Marinka},
journal={arXiv:2502.03569},
year={2025}
}
Code Availability
Pytorch implementation of CLEF is available in the GitHub repository.