Medical foundation models rely on tokenizing electronic health records (EHRs) into discrete sequences, but existing tokenizers treat medical codes as isolated text tokens, ignoring their structured relationships. With over 600,000 codes spanning diagnoses, treatments, and medications, capturing these dependencies is essential for accurate clinical reasoning.
We introduce MedTok, a multimodal medical code tokenizer that integrates both text descriptions and relational context. MedTok encodes medical codes using a language model for textual meaning and a graph encoder for hierarchical structure, unifying both into a shared token space. This approach ensures that medical codes are represented with richer, more context-aware embeddings.
Replacing standard tokenizers with MedTok significantly improves predictive performance across five EHR models, increasing AUPRC by 4.10% on MIMIC-III, 4.78% on MIMIC-IV, and 11.30% on EHRShot, with the largest gains in drug recommendation. Beyond EHR modeling, MedTok enhances medical QA systems, demonstrating its potential as a unified tokenizer for medical foundation models.
Motivation
Medical codes are the foundation of structured electronic health records (EHRs), encoding diagnoses, treatments, medications, and procedures in a standardized format. However, existing tokenization strategies treat these codes as isolated text tokens, disregarding their hierarchical relationships, co-occurrences, and domain-specific dependencies. With over 600,000 unique codes spanning multiple coding systems like ICD-10, SNOMED CT, and ATC, conventional tokenization leads to inefficient vocabulary expansion, loss of relational context, and fragmented representations that hinder medical AI models. These limitations affect the performance of foundation models trained on EHR data, limiting their ability to make accurate predictions, generate meaningful representations, and generalize across clinical tasks.
MedTok addresses this challenge by introducing a multimodal tokenizer that integrates both textual descriptions and relational knowledge from medical ontologies. Unlike traditional approaches that rely solely on text-based embeddings, MedTok combines language models with graph-based encodings to preserve the structure and semantics of medical codes. This unified tokenization framework enables foundation models to leverage both linguistic and ontological context, leading to more expressive token representations.
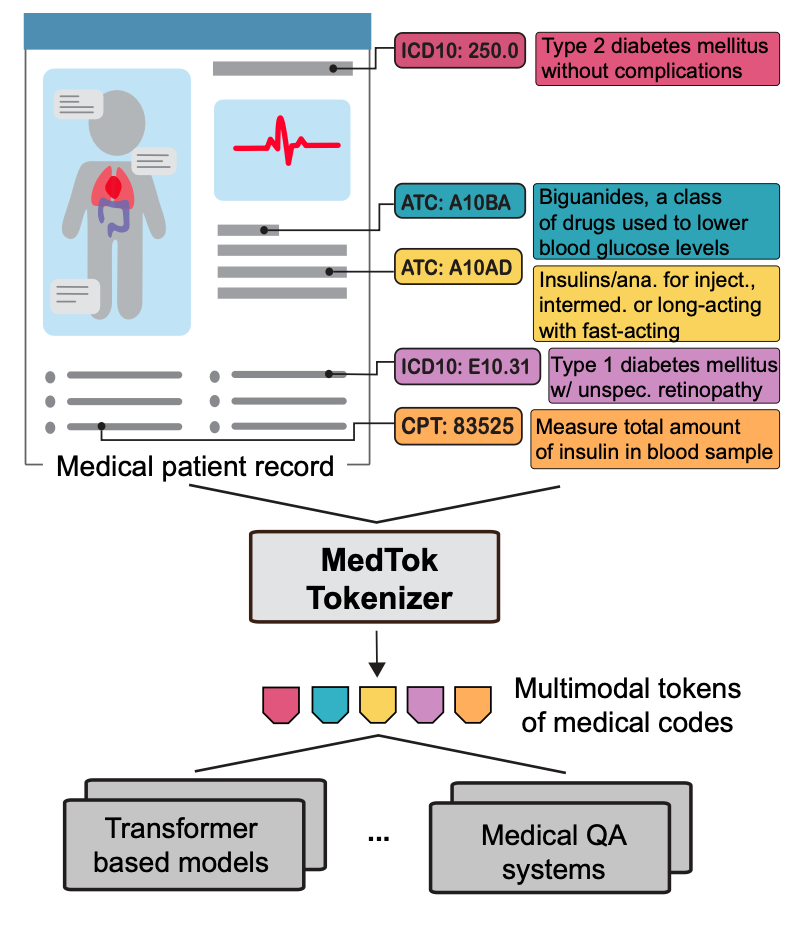
Overview of MedTok
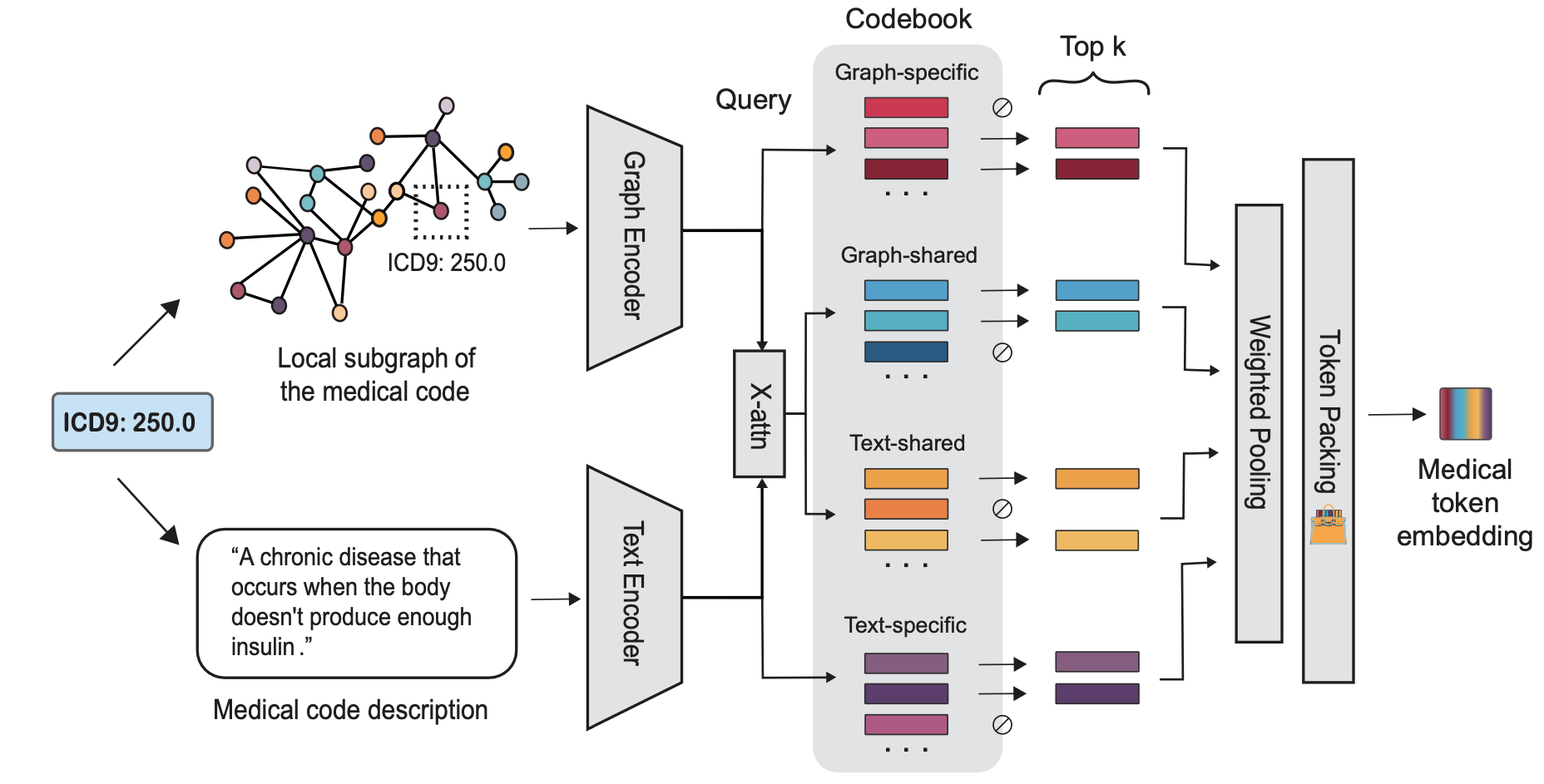
MedTok is a multimodal tokenizer designed to transform medical codes into structured token representations that preserve both textual semantics and hierarchical relationships. It achieves this by combining a text encoder, which processes the natural language descriptions of medical codes, with a graph encoder, which captures the dependencies and structured relations among codes. By jointly optimizing these two modalities, MedTok generates token embeddings that are both semantically rich and context-aware, allowing AI models to better interpret and utilize medical data.
MedTok operates through a three-step process:
- First, it encodes the textual descriptions of medical codes using a pre-trained language model, extracting meaningful embeddings.
- Next, it integrates graph-based representations from biomedical knowledge graphs, capturing relationships such as disease co-occurrence, drug interactions, and procedural hierarchies.
- Finally, it applies vector quantization to map both modalities into a shared token space, ensuring that information is retained in a format compatible with transformer-based models.
This process allows MedTok to serve as a drop-in replacement for existing tokenizers, improving model performance across a wide range of EHR-based applications.
MedTok tokenizer integrated with in-patient EHR transformer models
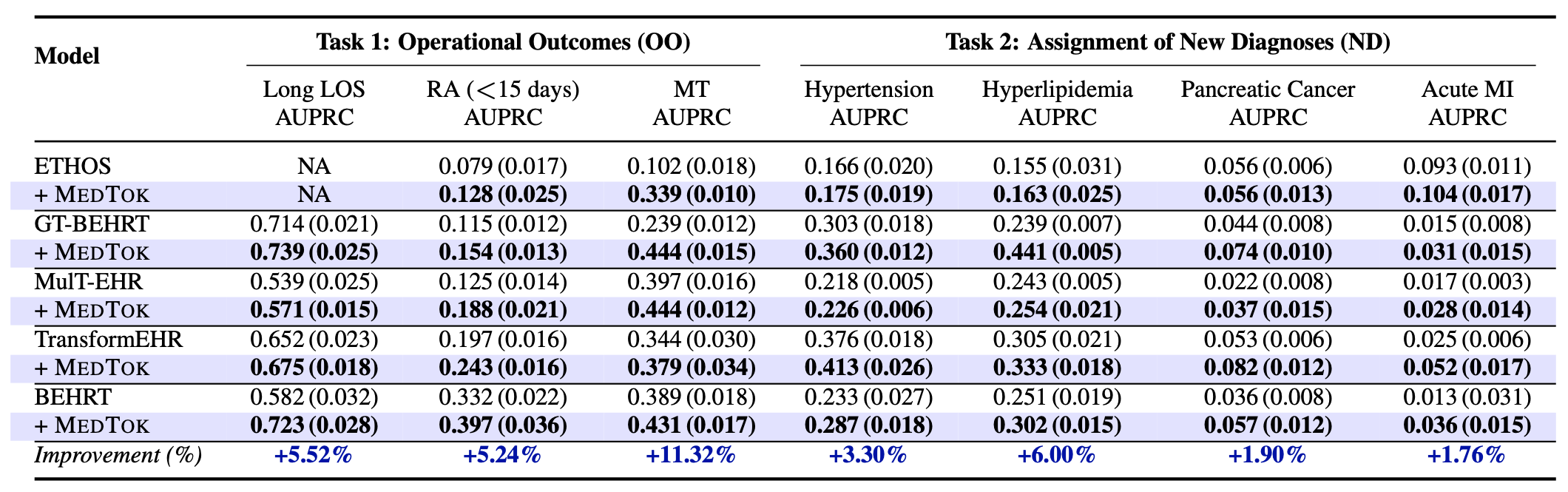
In the inpatient setting, MedTok is integrated with predictive models trained on ICU patient records from MIMIC-III and MIMIC-IV datasets. These datasets contain structured medical codes representing diagnoses, treatments, and lab results, which are critical for predicting clinical outcomes such as mortality risk, readmission likelihood, and length of stay. Standard tokenization approaches fail to capture dependencies between medical codes, leading to suboptimal performance in these predictive tasks.
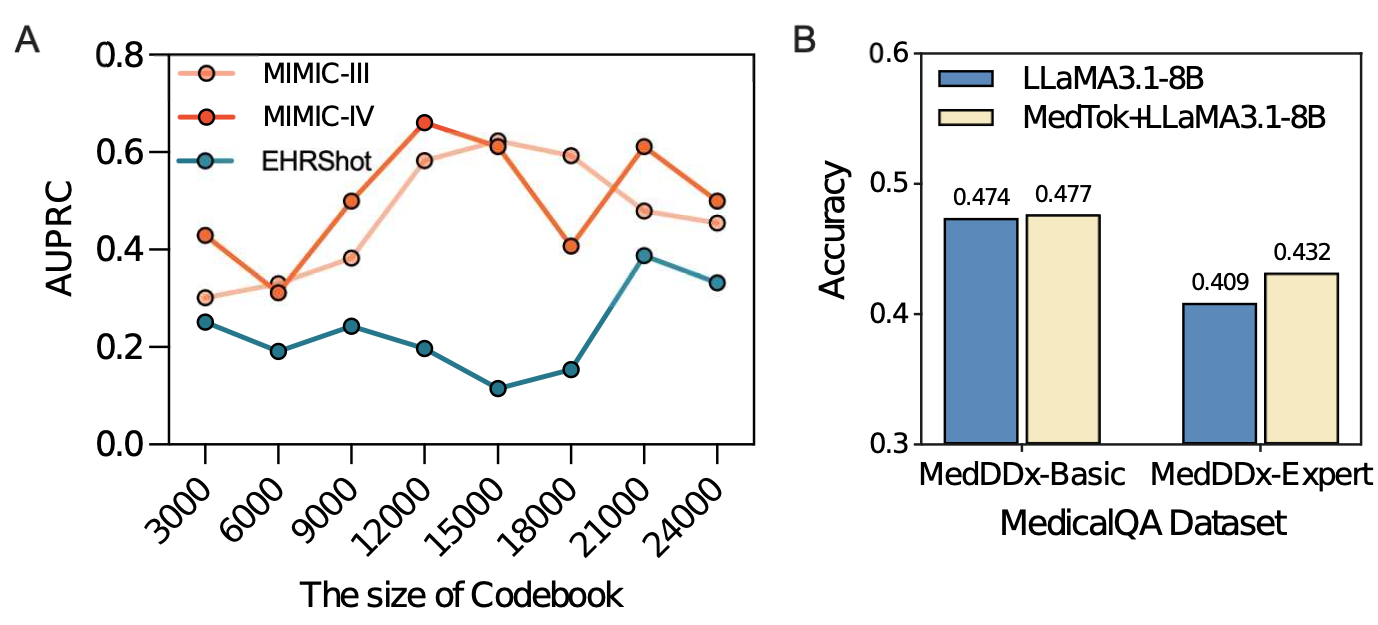
By replacing traditional tokenization methods with MedTok, we observe significant improvements across all clinical tasks. MedTok enhances AUPRC scores for mortality prediction, readmission prediction, and phenotype classification by up to 4.10% in MIMIC-III and 4.78% in MIMIC-IV. The largest gains are seen in drug recommendation tasks, where capturing the relationships between medications and diagnoses is crucial. These results demonstrate that MedTok enables more structured and context-aware tokenization, allowing transformer-based models to better understand inpatient EHR data.

MedTok tokenizer integrated with out-patient EHR transformer models
MedTok is also evaluated in outpatient settings using the EHRShot dataset, which includes longitudinal patient records spanning multiple visits across different healthcare facilities. Unlike inpatient datasets, outpatient records contain more diverse sequences of medical events, making it essential to capture long-term dependencies between medical codes. Standard tokenization methods often struggle with this variability, leading to inconsistencies in predictive performance.
Integrating MedTok into outpatient models results in substantial improvements in both operational and clinical tasks. The most significant impact is observed in mortality prediction, where MedTok increases AUPRC by 11.30%, demonstrating its ability to better capture disease trajectories over time. MedTok also improves new disease diagnosis assignments, particularly in detecting conditions such as acute myocardial infarction (8.80% improvement).
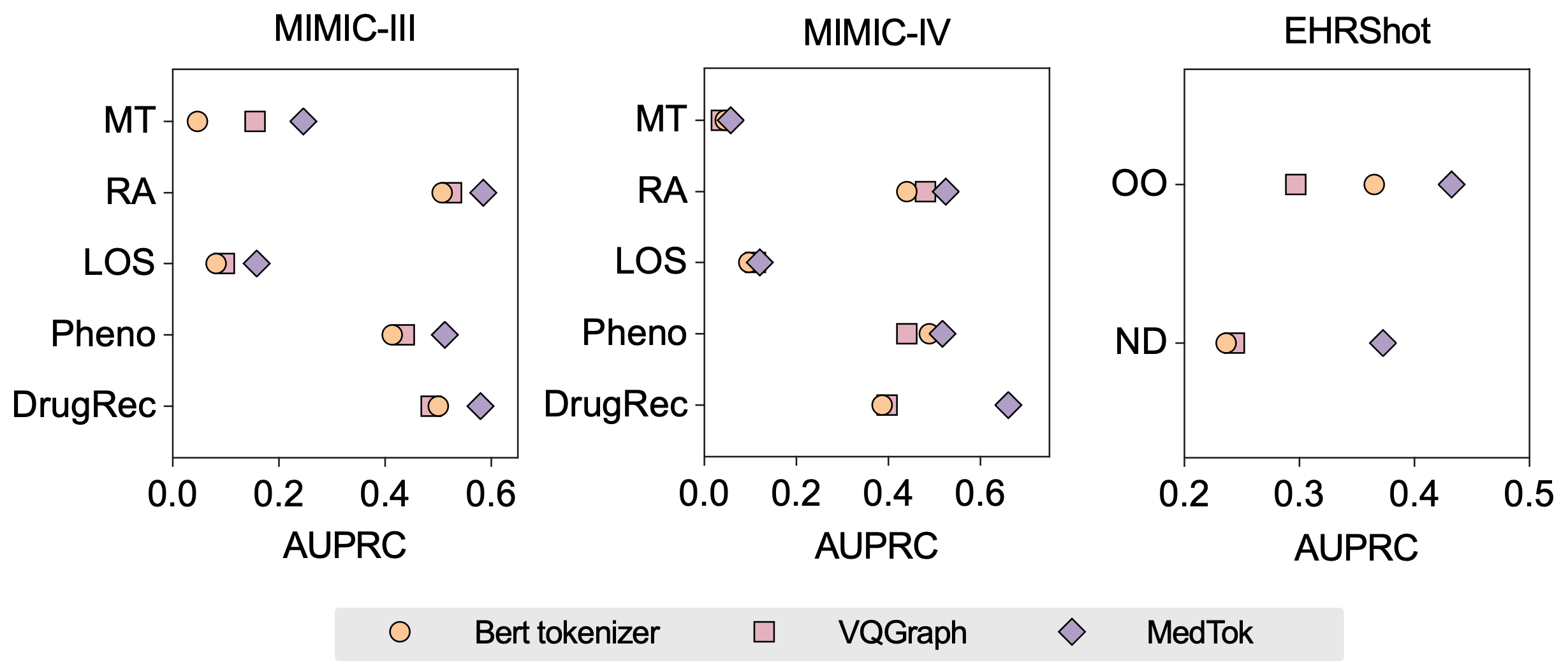
MedTok and alternative tokenization strategies
Standard tokenization methods in EHR modeling either treat medical codes as isolated text tokens or rely on manually curated embeddings that lack adaptability. Traditional text-based tokenizers, such as BERT-based models, struggle with the sheer scale of medical vocabularies and fail to capture relational dependencies between codes. Graph-based tokenizers offer some advantages by incorporating structured relationships but often lose important textual semantics in the process.
MedTok bridges this gap by integrating both modalities, ensuring that token representations retain the advantages of text-based semantics while embedding relational structures from medical ontologies. Compared to alternative tokenization strategies, MedTok consistently outperforms both text-only and graph-only approaches. Experimental results show that MedTok achieves higher AUPRC scores across inpatient and outpatient tasks, outperforming text-based tokenizers by up to 5.24% and graph-based tokenizers by up to 6.00%.
Benefits of multimodal tokenization
Ablation studies confirm the importance of both modalities in MedTok’s design. Removing the graph-based component leads to a decline in performance on drug recommendation and disease co-occurrence tasks, while removing the text-based component degrades performance in readmission prediction and phenotype classification.

MedTok is a general purpose tokenizer: Medical QA systems
Beyond EHR-based tasks, MedTok demonstrates its versatility by improving performance in medical question-answering (QA) systems. Many medical QA models rely on large language models trained on free-text clinical data but lack structured knowledge from medical coding systems.
We evaluate MedTok’s impact on the MedDDx dataset, which contains multiple-choice medical questions at different difficulty levels. Integrating MedTok with an LLM (LLaMA3.1-8B) leads to a 2.3% accuracy improvement on expert-level medical questions, demonstrating its ability to provide structured context for clinical reasoning.
Publication
Multimodal Medical Code Tokenizer
Xiaorui Su, Shvat Messica, Yepeng Huang, Ruth Johnson, Lukas Fesser, Shanghua Gao, Faryad Sahneh, Marinka Zitnik
International Conference on Machine Learning, ICML 2025 [arXiv]
@article{su2025multimodal,
title={Multimodal Medical Code Tokenizer},
author={Su, Xiaorui and Messica, Shvat and Huang, Yepeng and Johnson, Ruth and Fesser, Lukas and Gao, Shanghua and Sahneh, Faryad and Zitnik, Marinka},
journal={International Conference on Machine Learning, ICML},
year={2025}
}
Code Availability
MedTok is available on GitHub and model weights are hosted on HuggingFace.