We establish a key connection between counterfactual fairness and stability and leverage it to develop NIFTY (uNIfying Fairness and stabiliTY), a novel framework that can be used with any GNN to learn fair and stable representations.
We introduce a novel objective function that simultaneously accounts for fairness and stability and develop a layer-wise weight normalization using the Lipschitz constant to enhance neural message passing in GNNs. In doing so, we enforce fairness and stability both in the objective function as well as in the GNN architecture. Further, we show theoretically that our layer-wise weight normalization promotes counterfactual fairness and stability in the resulting representations.
We introduce three new graph datasets comprising of high-stakes decisions in criminal justice and financial lending domains. Extensive experimentation with the above datasets demonstrates the efficacy of our framework.
Publication
Towards a Unified Framework for Fair and Stable Graph Representation Learning
Chirag Agarwal, Himabindu Lakkaraju*, Marinka Zitnik*
Conference on Uncertainty in Artificial Intelligence, UAI 2021 [arXiv] [poster] [ICML 2021 Socially Responsible ML]
@inproceedings{agarwal2021towards,
title={Towards a Unified Framework for Fair and Stable Graph Representation Learning},
author={Agarwal, Chirag and Lakkaraju, Himabindu and Zitnik, Marinka},
booktitle={Proceedings of Conference on Uncertainty in Artificial Intelligence, UAI},
year={2021}
}
Motivation
Over the past decade, there has been a surge of interest in leveraging GNNs for graph representation learning. GNNs have been used to learn powerful representations that enabled critical predictions in downstream applications—e.g., predicting protein-protein interactions, drug repurposing, crime forecasting, news and product recommendations.
As GNNs are increasingly implemented in real-world applications, it becomes important to ensure that these models and the resulting representations are safe and reliable. More specifically, it is important to ensure that:
- these models and the representations they produce are not perpetrating undesirable discriminatory biases (i.e., they are fair), and
- these models and the representations they produce are robust to attacks resulting from small perturbations to the graph structure and node attributes (i.e., they are stable).
NIFTY framework
We first identify a key connection between counterfactual fairness and stability. While stability accounts for robustness w.r.t. small random perturbations to node attributes and/or edges, counterfactual fairness accounts for robustness w.r.t. modifications of the sensitive attribute.
We leverage this connection to develop NIFTY that can be used with any existing GNN model to learn fair and stable representations. Our framework exploits the aforementioned connection to enforce fairness and stability both in the objective function as well as in the GNN architecture.
More specifically, we introduce a novel objective function which simultaneously optimizes for counterfactual fairness and stability by maximizing the similarity between representations of the original nodes in the graph, and their counterparts in the augmented graph. Nodes in the augmented graph are generated by slightly perturbing the original node attributes and edges or by considering counterfactuals of the original nodes where the value of the sensitive attribute is modified. We also develop a novel method for improving neural message passing by carrying out layer-wise weight normalization using the Lipschitz constant.
We theoretically show that this normalization promotes counterfactual fairness and stability of learned representations. To the best of our knowledge, this work is the first to tackle the problem of learning node representations that are both fair and stable.
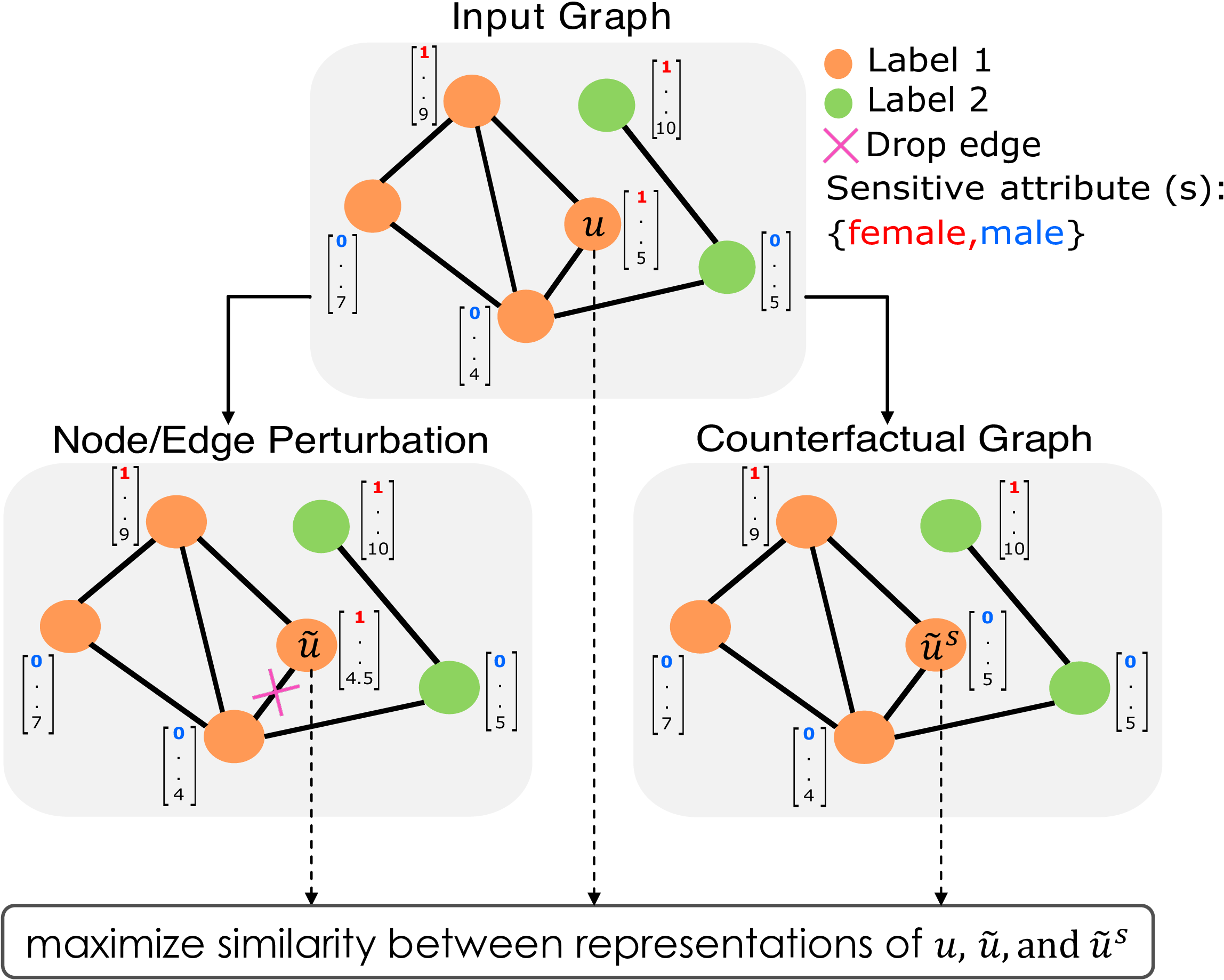
The figure above gives an overview of NIFTY. NIFTY can learn node representations that are both fair and stable (i.e., invariant to the sensitive attribute value and perturbations to the graph structure and non-sensitive attributes) by maximizing the similarity between representations from diverse augmented graphs.
Datasets
We introduce and experiment with three new graph datasets comprising of critical decisions in criminal justice (if a defendant should be released on bail) and financial lending (if an individual should be given loan) domains.
- German credit graph has 1,000 nodes representing clients in a German bank that are connected based on the similarity of their credit accounts. The task is to classify clients into good vs. bad credit risks considering clients’ gender as the sensitive attribute.
- Recidivism graph has 18,876 nodes representing defendants who got released on bail at the U.S state courts during 1990-2009. Defendants are connected based on the similarity of past criminal records and demographics. The task is to classify defendants into bail (i.e., unlikely to commit a violent crime if released) vs. no bail (i.e., likely to commit a violent crime) considering race information as the protected attribute.
- Credit defaulter graph has 30,000 nodes representing individuals that we connected based on the similarity of their spending and payment patterns. The task is to predict whether an individual will default on the credit card payment or not while considering age as the sensitive attribute.
Code
Source code is available in the GitHub repository.