Adverse events from medications accounted for over 110,000 deaths in the US alone in 2019. The impact of the COVID-19 pandemic on patient safety and how it exacerbated pre-existing inequalities across diverse patient cohorts holds urgent clinical questions. However, intricate dependencies between the pandemic’s effects, safety profiles of drugs, and patient characteristics pose challenges to extracting clinically actionable insights.
An algorithmic approach to investigate the impact of COVID-19 on adverse drug safety and identify at risk cohorts is missing from the literature. In this project, we develop a model to reveal impacts of the pandemic on drug safety and identify at risk demographics from 10,443,476 adverse event reports spanning 19,193 adverse events and 3,624 drugs collected by the FDA Adverse Event Reporting System (FAERS).
Urgent need for algorithms aimed at improving safe medication use
Adverse events from medications accounted for over 110,000 deaths in the U.S. alone in 2019. The pandemic has further challenged healthcare systems’ ability to ensure safe medication use. Despite these urgent implications, the pandemic’s effects on adverse drugs effects remain unknown. Further, intricate dependencies between the pandemic’s effects, drugs, and patient characteristics present unique challenges for understanding patient safety during a public health emergency:
a) Algorithmic approaches are needed to unveil how the patient safety landscape changed with the pandemic onset. Such studies would reveal what inequalities in patient populations are exacerbated more than expected had the pandemic not occurred.
b) Algorithmic approaches are needed to compare patient safety to its pre-pandemic levels across patient groups and the entire range of human diseases and approved drugs.
Addressing this challenge can (i) inform drug prescription, (ii) improve patient safety by identifying individuals at high risk for adverse events and those who are disproportionally affected by preventable inequities, and (iii) enable comparison of COVID-19 pandemic to other health emergencies to unveil the disruptive nature of public health crises and inform health policy.
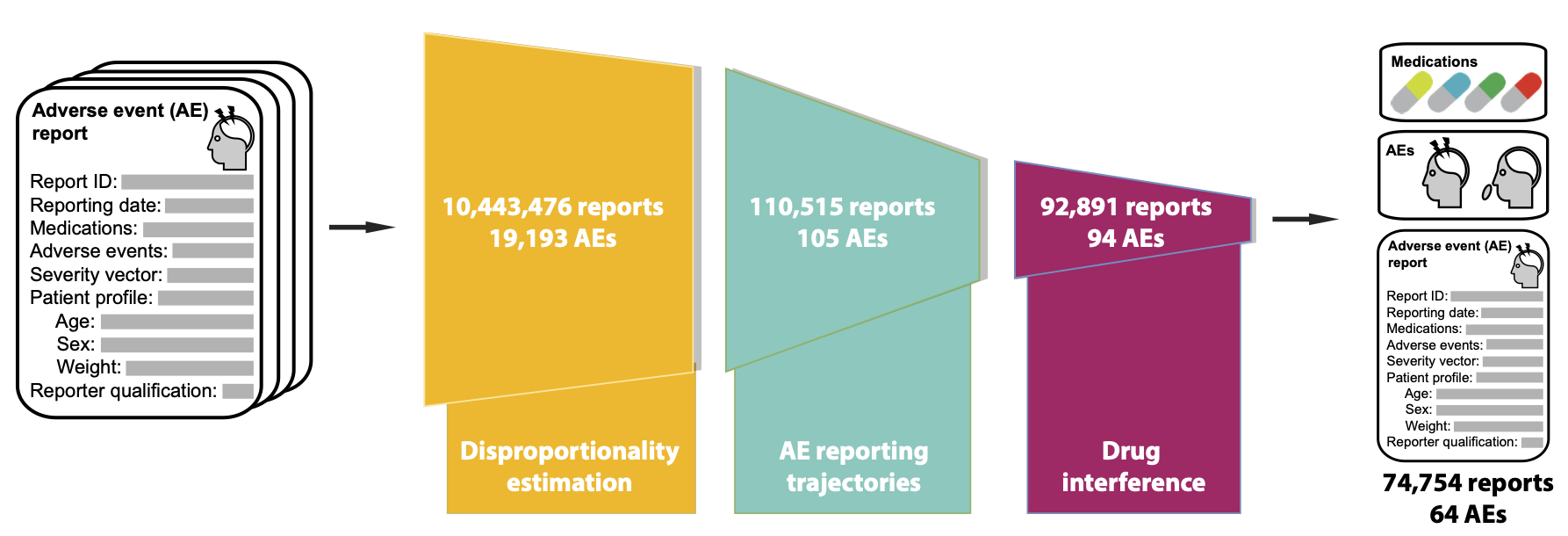
Novel algorithmic approach to drug safety
This study addresses the above challenge, representing the first study to unveil the disruptive nature of a public health crisis on patient safety. Using the largest dataset so far, consisting of 10,443,476 adverse drug event reports spanning 7 years (Jan. 2013-Sept 2020) and involving 3,624 drugs and 19,193 adverse events, we develop an algorithmic approach to investigate negative outcomes associated with medication use and how they changed during the pandemic.
In contrast to previous methods focused on small subsets of the adverse event landscape, our model investigates the entire range of human diseases and approved drugs. Our approach contains three key components: identifying adverse events whose incidence has significantly changed after the pandemic, removing temporal confounding factors in reporting trajectories, and pinpointing adverse events with considerable associations to medications.
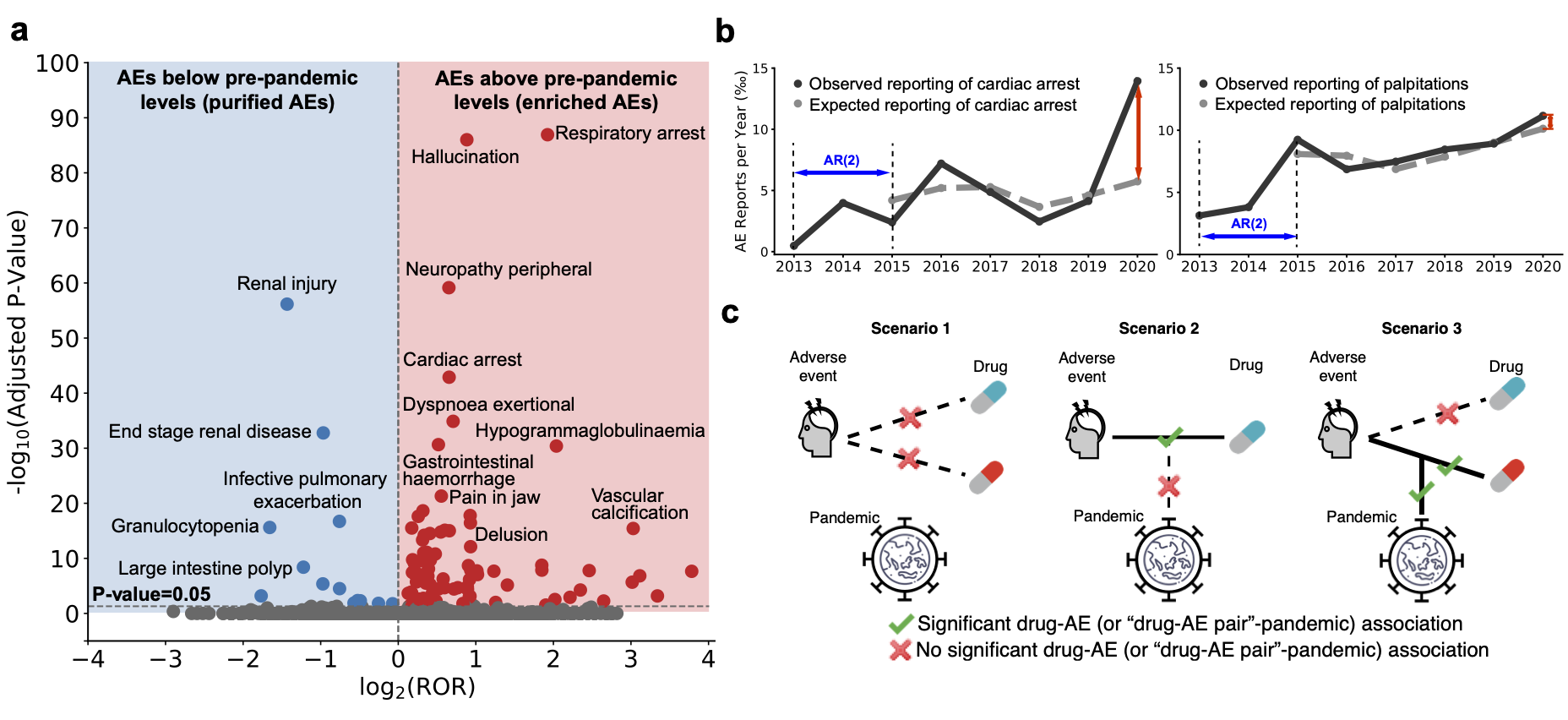
Advantages of our algorithmic approach
1) Detect subtle patterns of drug safety: By correcting for confounding factors like temporal reporting trajectories, our model can detect impacts of the pandemic even in rare adverse events.
2) Generalizable: Our three step approach can flexibly identify differential reporting patterns in patient cohorts formed as a function of gender, age, adverse events, and drug.
3) New resource of adverse events and drug-event associations: This resource can be readily applied for use in pharmacoepidemiology and public health policy to inform medication use in diverse populations.
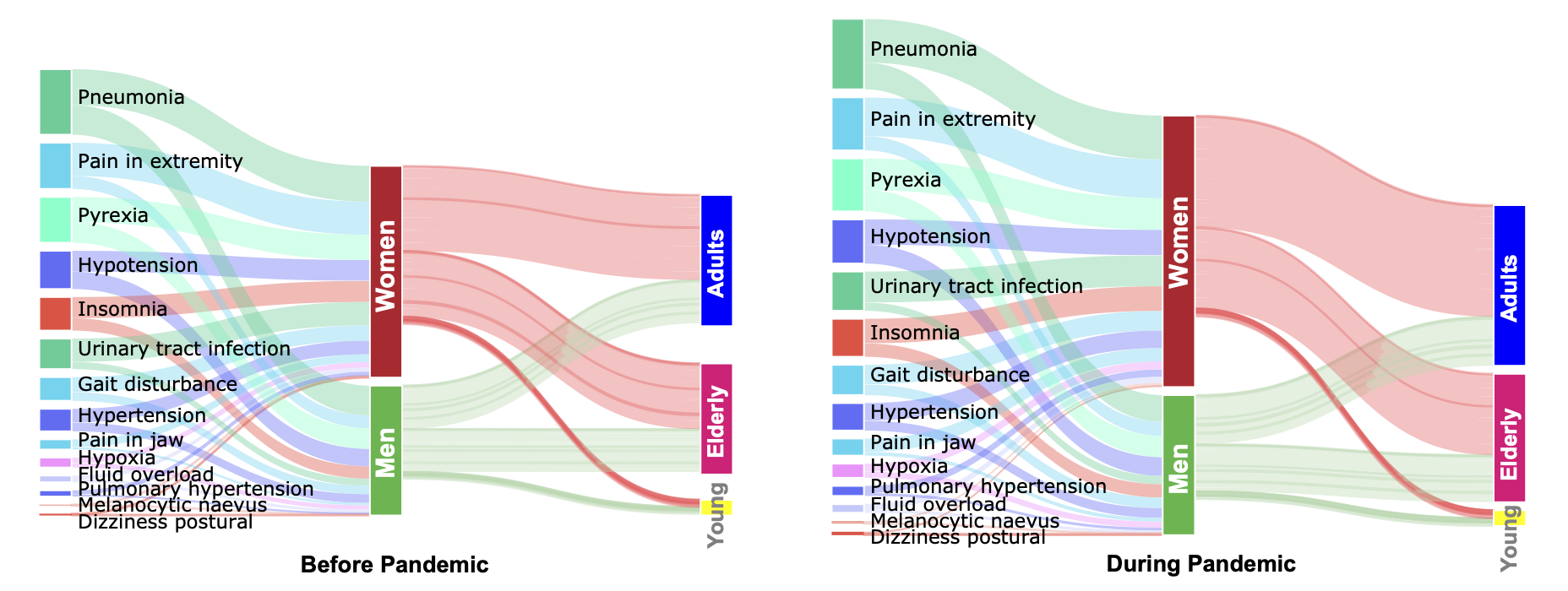
Key results and findings
Our algorithmic effort leads to several key findings:
-
We find substantial variation in adverse drug events before and during the pandemic. Among 64 adverse events identified by our analyses, we find 54 have increased incidence rates during the pandemic, even though adverse event reporting decreased by 4.4% overall.
-
We find that pre-pandemic gender differences are exaggerated during the pandemic. Women suffer from more drug adverse events than men relative to pre-pandemic levels, across all age cohorts. Comparing to male patients, women report 47.0% more adverse events whose occurrence significantly increased during the pandemic relative to pre-pandemic levels. Out of 53 adverse events with the pre-pandemic gender gap, 33 have increased gap during the pandemic more than expected had the pandemic not occurred.
-
We also find relevant clinical differences in adverse drug events outcomes across age groups. For example, acute kidney injury has seen a surprising increase in adult but not in elderly patients, suggesting an age-related disparity in the pandemic’s impact on drug-related kidney injury. In contrast, mental health-related events (hallucination, delusion, aggression, abnormal behavior, and dementia) have disproportionately increased in women and the elderly, indicating they constitute at risk patient cohorts. In contrast to narrowly focused prior studies, our work can unmix a population to identify patients at higher risk for adverse events during the pandemic than in the pre-pandemic time.
-
The algorithmic approach also identifies how the impact of adverse drug reactions on human organs changed during the pandemic. For example, while musculoskeletal and metabolic side effects are disproportionately found in women during the pandemic, immune- related adverse events are enriched only in men.
-
Detection of rare adverse events for Remdesivir. We detect novel rare adverse events such as hypoxia for Remdesivir, highlighting the role for algorithmic models for medications granted emergency approval.
-
Finally, we present a new resource of adverse drug effects and drug-event associations for use in pharmacoepidemiology and public health policy to inform safe medication use.
Broad impact
Our findings have implications for safe medication use and highlight the role of variation in adverse events for improving patient safety during a public health emergency.
a) Our algorithmic approach can identify differential reporting patterns in patient cohorts formed as a function of gender, age, adverse events, and drugs. With additional information on medical and non-medical characteristics, the approach is suitable for systematic safety surveillance to pinpoint individuals at high risk for safety events based on risk-altering interactions.
b) We expect this algorithmic approach to enable comparison of the COVID-19 pandemic to other health emergencies (like the nationwide opioid crisis in the U.S. and emergencies resulting hurricanes and wildfires) to unveil the disruptive nature of public health crises on patient safety.
c) Finally, our research can inform safe medication use by identifying populations at high risk for adverse events and proactive vigilance of vaccine programs in large and diverse populations.
Publication
Population-Scale Identification of Differential Adverse Events Before and During a Pandemic
Xiang Zhang, Marissa Sumathipala, Marinka Zitnik
Nature Computational Science 2021
@article{zhang2021population,
title={Population-scale identification of differential adverse events before and during a pandemic},
author={Zhang, Xiang and Sumathipala, Marissa and Zitnik, Marinka},
journal={Nature Computational Science},
volume={1},
number={10},
pages={666--677},
year={2021},
publisher={Nature Publishing Group}
}
Code
Python implementation of the methodology developed and used in this project is available via GitHub repository.
Datasets
All data used in the paper, including the raw and processed adverse event report dataset, adverse event ontology, drug ontology, the final and intermediate results of the analyses are shared with research community via Harvard Dataverse repository. The dataset is uniquely identified as https://doi.org/10.7910/DVN/G9SHDA.
Direct access to pre-processed datasets
Direct access to results
-
Associations_between_adverse_events_and_the_pandemic_Top100.csv
-
Associations_between_drug_adverse_event_pairs_and_the_pandemic.csv
-
Associations_between_drug_adverse_event_pairs_and_the_pandemic_Top100.csv