Interpreting time series models is uniquely challenging because it requires identifying both the location of time series signals that drive model predictions and their matching to an interpretable temporal pattern. While explainers from other modalities can be applied to time series, their inductive biases do not transfer well to the inherently uninterpretable nature of time series.
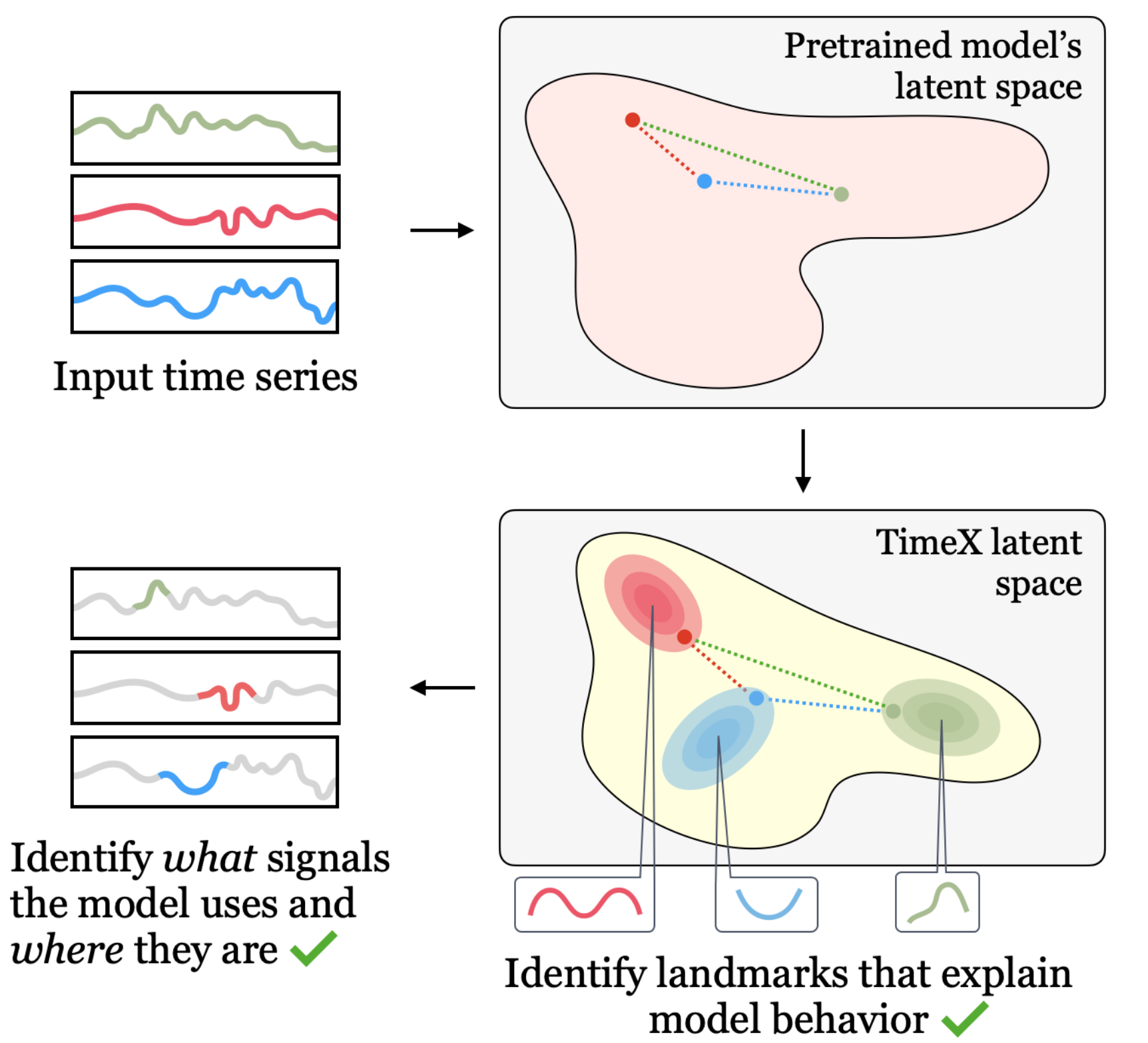
We present TimeX, a time series consistency model for training explainers. TimeX trains an interpretable surrogate to mimic the behavior of a pretrained time series model. It addresses the issue of model faithfulness by introducing model behavior consistency, a novel formulation that preserves relations in the latent space induced by the pretrained model with relations in the latent space induced by TimeX. TimeX provides discrete attribution maps and, unlike existing interpretability methods, it learns a latent space of explanations that can be used in various ways, such as to provide landmarks to visually aggregate similar explanations and easily recognize temporal patterns.
We evaluate TimeX on eight synthetic and real-world datasets and compare its performance against state-of-the-art interpretability methods. We also conduct case studies using physiological time series, showing that the novel components of TimeX show potential for training faithful, interpretable models that capture the behavior of pretrained time series models.
State-of-the-art time series models are high-capacity pre-trained neural networks often seen as black boxes due to their internal complexity and lack of interpretability. However, practical use requires techniques for auditing and interrogating these models to rationalize their predictions. Interpreting time series models poses a distinct set of challenges due to the need to achieve two goals:
- pinpointing the specific location of time series signals that influence the model’s predictions,
- aligning those signals with interpretable temporal patterns.
Research in model understanding and interpretability developed post-hoc explainers that treat pretrained models as black boxes and do not need access to internal model parameters, activations, and gradients. Recent research, however, shows that such post hoc methods suffer from a lack of faithfulness and stability, among other issues. A model can also be understood by investigating what parts of the input it attends to through attention mapping and measuring the impact of modifying individual computational steps within a model. Another major line of inquiry investigates internal mechanisms by asking what information the model contains. For example, it has been found that even when a language model is conditioned to output falsehoods, it may include a hidden state that represents the true answer internally. Such a gap between external failure modes and internal states can only be identified by probing model internals. Such representation probing has been used to characterize the behaviors of language models, but leveraging these strategies to understand time series models has yet to be attempted.
These lines of inquiry drive the development of in-hoc explainers that build inherent interpretability into the model through architectural modifications or regularization. However, no in-hoc explainers have been developed for time series data. While explainers designed for other modalities can be adapted to time series, their inherent biases translate poorly to the uninterpretable nature of time series data and can miss important structures in time series.
Explaining time series models is challenging for many reasons:
- Large time series data are not visually interpretable, as opposed to imaging or text datasets.
- Time series often exhibit dense informative features, in contrast to more explored modalities such as imaging, where informative features are often sparse. In time series datasets, timestep-to-timestep transitions can be negligible and temporal patterns only show up when looking at time segments and long-term trends. In contrast, in text datasets, word-to-word transitions are informative for language modeling and understanding. Time series interpretability involves understanding dynamics of the model and identifying trends or patterns.
- Another key issue with applying prior methods is that they treat all time steps as separate features, ignoring potential time dependencies and contextual information; we need explanations that are temporally connected and visually digestible.
- While understanding predictions of individual samples is valuable, the ability to establish connections between explanations of various samples (for example, in an appropriate latent space) could help alleviate these challenges.
Overview of TimeX
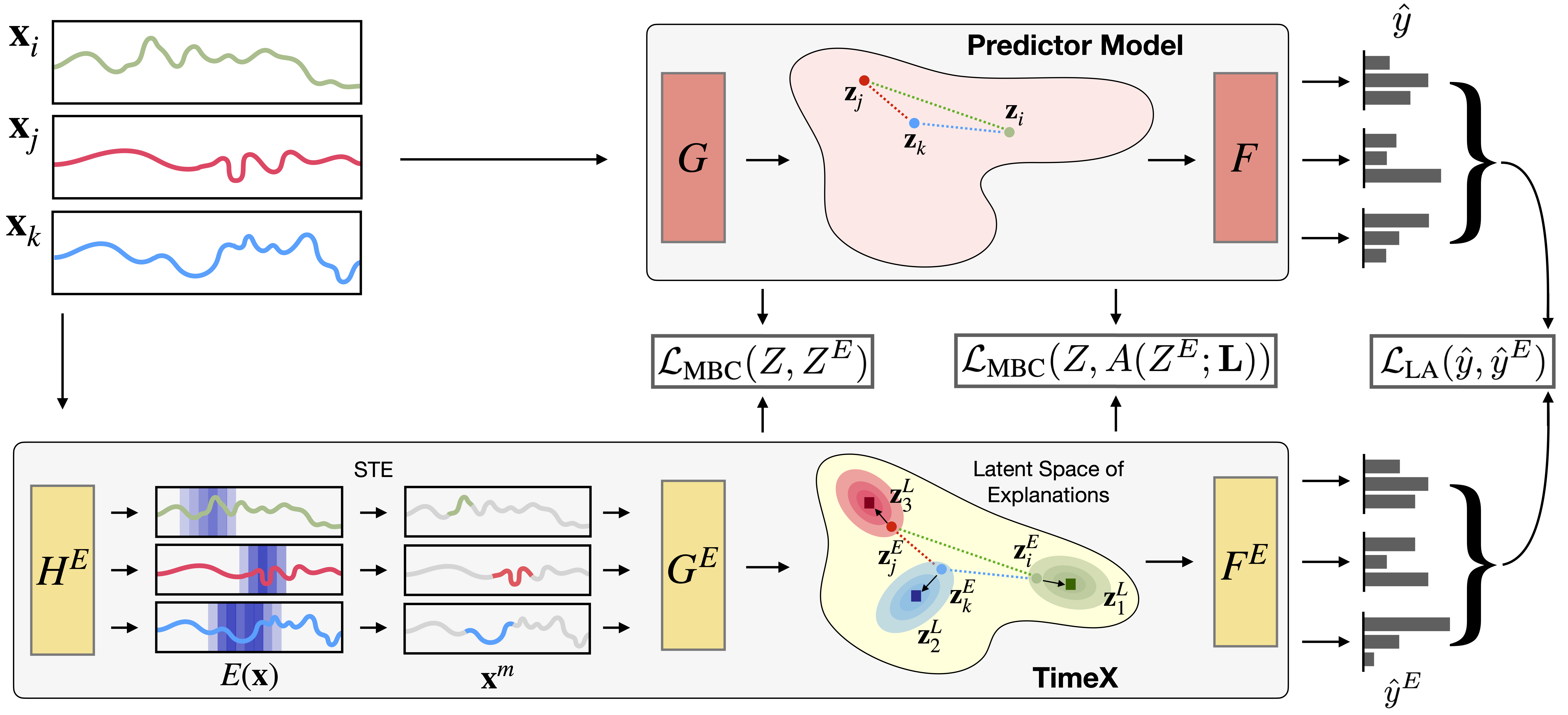
TimeX is a novel time series in-hoc explainer that produces interpretable attribution masks as explanations over time series inputs. An essential contribution of TimeX is the introduction of model behavior consistency, a novel formulation that ensures the preservation of relationships in the latent space induced by the pretrained model and the latent space induced by TimeX.
In addition to achieving model behavior consistency, TimeX offers interpretable attribution maps, which are valuable tools for interpreting the model’s predictions, generated using discrete straight-through estimators, a type of gradient estimator that enable end-to-end training of TimeX models.
Unlike existing interpretability methods, TimeX goes further by learning a latent space of explanations. By incorporating model behavior consistency and leveraging a latent space of explanations, TimeX provides discrete attribution maps and visual summaries of similar explanations with interpretable temporal patterns.
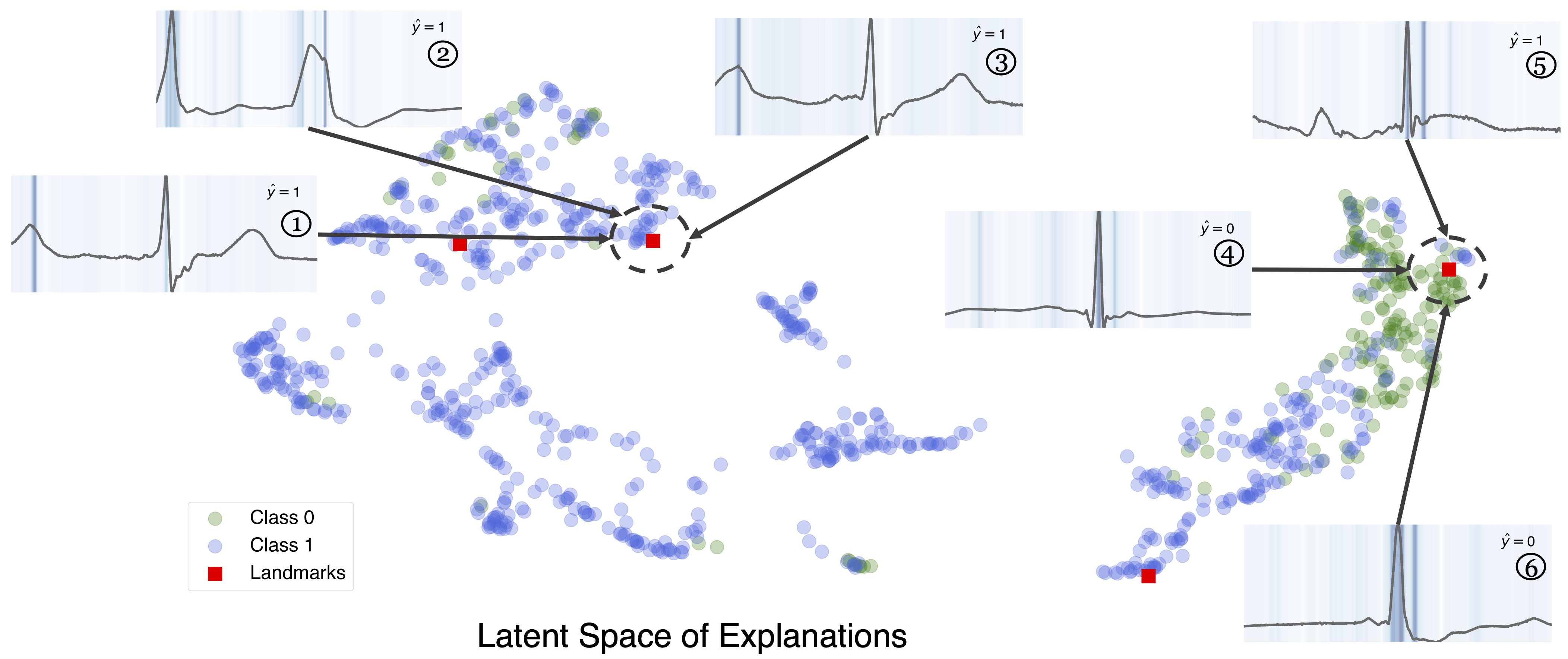
Landmark Explanation Analysis Using an ECG Dataset
To demonstrate TimeX’s landmarks, we show how landmarks serve as summaries of diverse patterns in the ECG dataset. The figure below visualizes the learned landmarks in the latent space of explanations. We choose four representative landmarks. Every landmark occupies different regions of the latent space, capturing diverse types of explanations generated by the model.
We show the three nearest explanations for the top two landmarks in terms of the nearest neighbor in the latent space. Explanations ①, ②, and ③ are all similar to each other while distinctly different from ④, ⑤, and ⑥, both in terms of attribution and temporal structure. This visualization shows how landmarks can partition the latent space of explanations into interpretable temporal patterns.
Publication
Encoding Time-Series Explanations through Self-Supervised Model Behavior Consistency
Owen Queen, Thomas Hartvigsen, Teddy Koker, Huan He, Theodoros Tsiligkaridis, Marinka Zitnik
Proceedings of Neural Information Processing Systems, NeurIPS 2023 [arXiv]
@inproceedings{queen2023encoding,
title = {Encoding Time-Series Explanations through Self-Supervised Model Behavior Consistency},
author = {Queen, Owen and Hartvigsen, Thomas and Koker, Teddy and Huan, He and Tsiligkaridis, Theodoros and Zitnik, Marinka},
booktitle = {Proceedings of Neural Information Processing Systems, NeurIPS},
year = {2023}
}
Code
Pytorch implementation of TimeX is available in the GitHub repository.