Developing personalized diagnostic strategies and targeted treatments requires a deep understanding of disease biology and the ability to dissect the relationship between molecular and genetic factors and their phenotypic consequences. However, such knowledge is fragmented across publications, non-standardized research repositories, and evolving ontologies describing various scales of biological organization between genotypes and clinical phenotypes.
We introduce PrimeKG, a precision medicine-oriented knowledge graph that provides a holistic view of diseases. PrimeKG integrates 20 high-quality resources to describe 17,080 diseases with 4,050,249 relationships representing ten major biological scales, including disease-associated protein perturbations, biological processes and pathways, anatomical and phenotypic scale, and the entire range of approved and experimental drugs with their therapeutic action, considerably expanding previous efforts in disease-rooted knowledge graphs.
PrimeKG supports drug-disease prediction by including an abundance of ’indications’, ’contradictions’ and ’off-label use’ edges, which are usually missing in other knowledge graphs. We accompany PrimeKG's graph structure with text descriptions of clinical guidelines for drugs and diseases to enable multi-modal analyses.
The figure below provides an overview of PrimeKG. Panel a shows a schematic overview of the various types of nodes in PrimeKG and the relationships they have with other nodes in the graph.
Panel b shows all disease nodes in PrimeKG visualized in a circular layout together with disease-associated information. Shown are relationships between disease nodes and any other node type. Disease nodes are densely connected to four other node types in PrimeKG through seven types of relations.
Panel c shows an example of paths in PrimeKG between the disease node ‘Autism’ and the drug node ‘Risperidone’. Intermediate nodes are colored by their node type from panel a. We also display snippets of text features for both nodes to demonstrate the multimodal nature of PrimeKG.
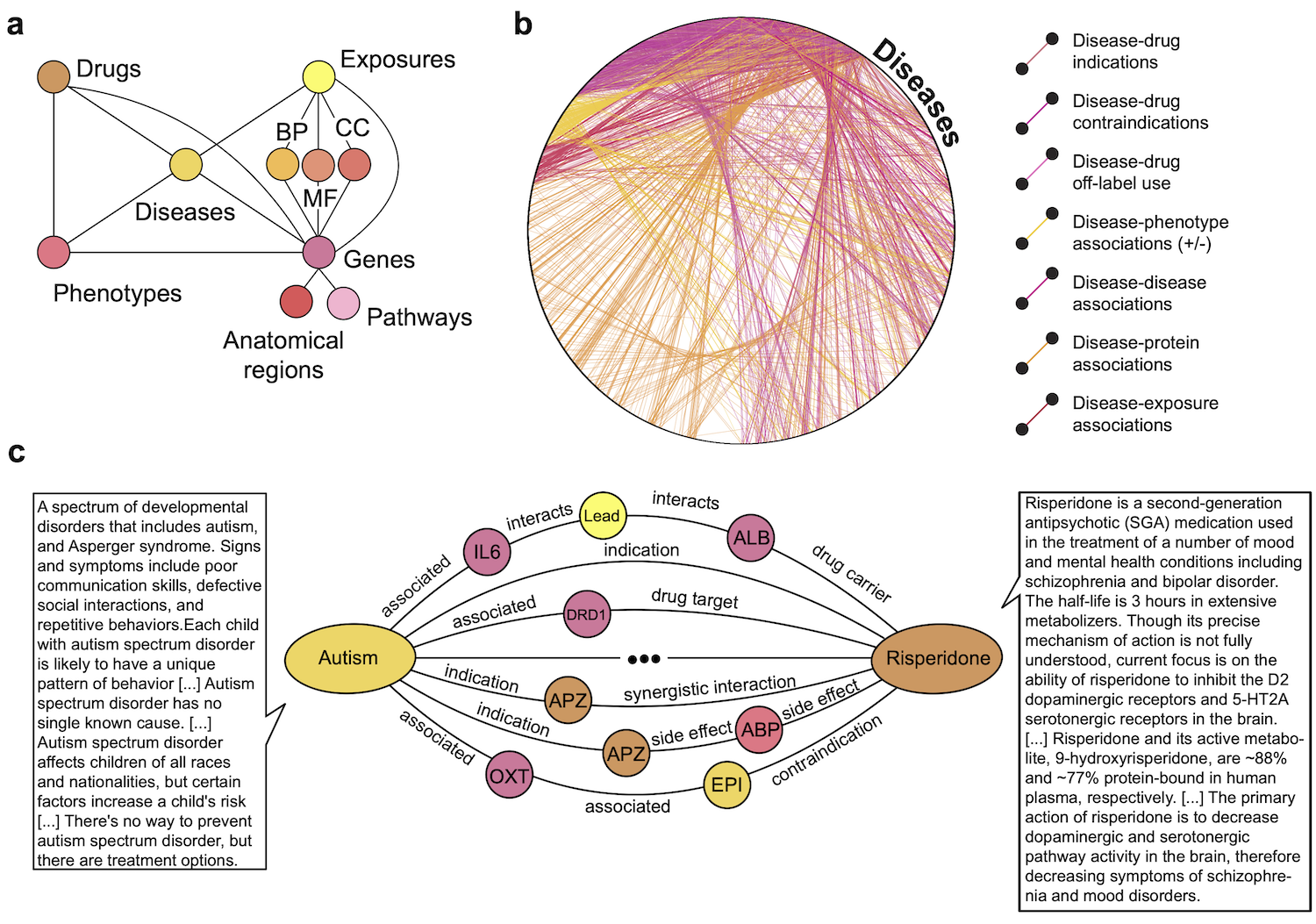
Abbreviations - MF: molecular function, BP: biological process, CC: cellular component, APZ: Apiprazole, EPI: epilepsy, ABP: abdominal pain, + / - associations: positive and negative associations.
Publication
Building a knowledge graph to enable precision medicine
Payal Chandak*, Kexin Huang*, and Marinka Zitnik
Scientific Data 2023 [bioRxiv]
@article{chandak2023building,
title={Building a knowledge graph to enable precision medicine},
author={Chandak, Payal and Huang, Kexin and Zitnik, Marinka},
journal={Scientific Data},
volume={10},
number={1},
pages={67},
url={https://doi.org/10.1038/s41597-023-01960-3},
year={2023},
publisher={Nature Publishing Group}
}
Code
The code to reproduce results, together with documentation and tutorials, is available in PrimeKG’s Github repository.
Data availability
PrimeKG is hosted on Harvard Dataverse. We deposited the knowledge graph along with all relevant intermediate files at this repository.