Protein interaction networks are a critical component in studying the function and therapeutic potential of proteins. However, accurately modeling protein interactions across diverse biological contexts, such as tissues and cell types, remains a significant challenge for existing algorithms.
We introduce PINNACLE, a flexible geometric deep learning approach that is trained on contextualized protein interaction networks to generate context-PINNACLE protein representations. Leveraging a human multi-organ single-cell transcriptomic atlas, PINNACLE provides 394,760 protein representations split across 156 cell type contexts from 24 tissues and organs.
We demonstrate that PINNACLE's contextualized representations of proteins reflect cellular and tissue organization and PINNACLE's tissue representations enable zero-shot retrieval of the tissue hierarchy. Infused with cellular and tissue contexts, PINNACLE's protein representations can be adapted for downstream tasks: to enhance 3D structure-based protein representations (namely, PD-1/PD-L1 and B7-1/CTLA-4) and to study the genomic effects of drugs across cellular contexts. Enabled by contextualized learning, PINNACLE's protein representations outperform state-of-the-art, yet context-free, models in nominating therapeutic targets for rheumatoid arthritis and inflammatory bowel diseases in at least 18.6% (29 out of 156) and 8.6% (13 out of 152) of cell type contexts, respectively. PINNACLE empowers the long-standing paradigm of incorporating biological context into artificial intelligence models to better model biological systems.
Proteins are the functional units of cells, and their interactions allow performing different biological functions. The development of high-throughput methods has enabled the characterization of large maps of protein interactions. Leveraging these protein interaction networks, computational methods have been developed to improve the understanding of protein structure, accurately predict functional annotations, and inform the design of therapeutic targets.
The roles of proteins are influenced by the biological contexts in which they act:
- Proteins can have distinct roles in different contexts. While nearly every cell contains the same genome, the expression of genes and the function of proteins encoded by these genes depend on cellular and tissue contexts. Gene expression and the function of proteins can also differ significantly between healthy and disease states. Therefore, computational methods that incorporate biological contexts can improve the characterization of proteins.
- Existing methods produce protein representations (or embeddings) that are context-free. Each protein has only one representation learned from either a single context or an integrated view across many contexts. These methods generate one representation for each protein, providing an integrated summary of the protein. While such protein representations can be valuable, they are not tailored to specific biological contexts, such as cell types and disease states. This makes it challenging to use protein representations for predicting molecular phenotypes that vary with cell types as well as for predicting pleiotropy and other protein roles in distinct cell types.
Overview of PINNACLE
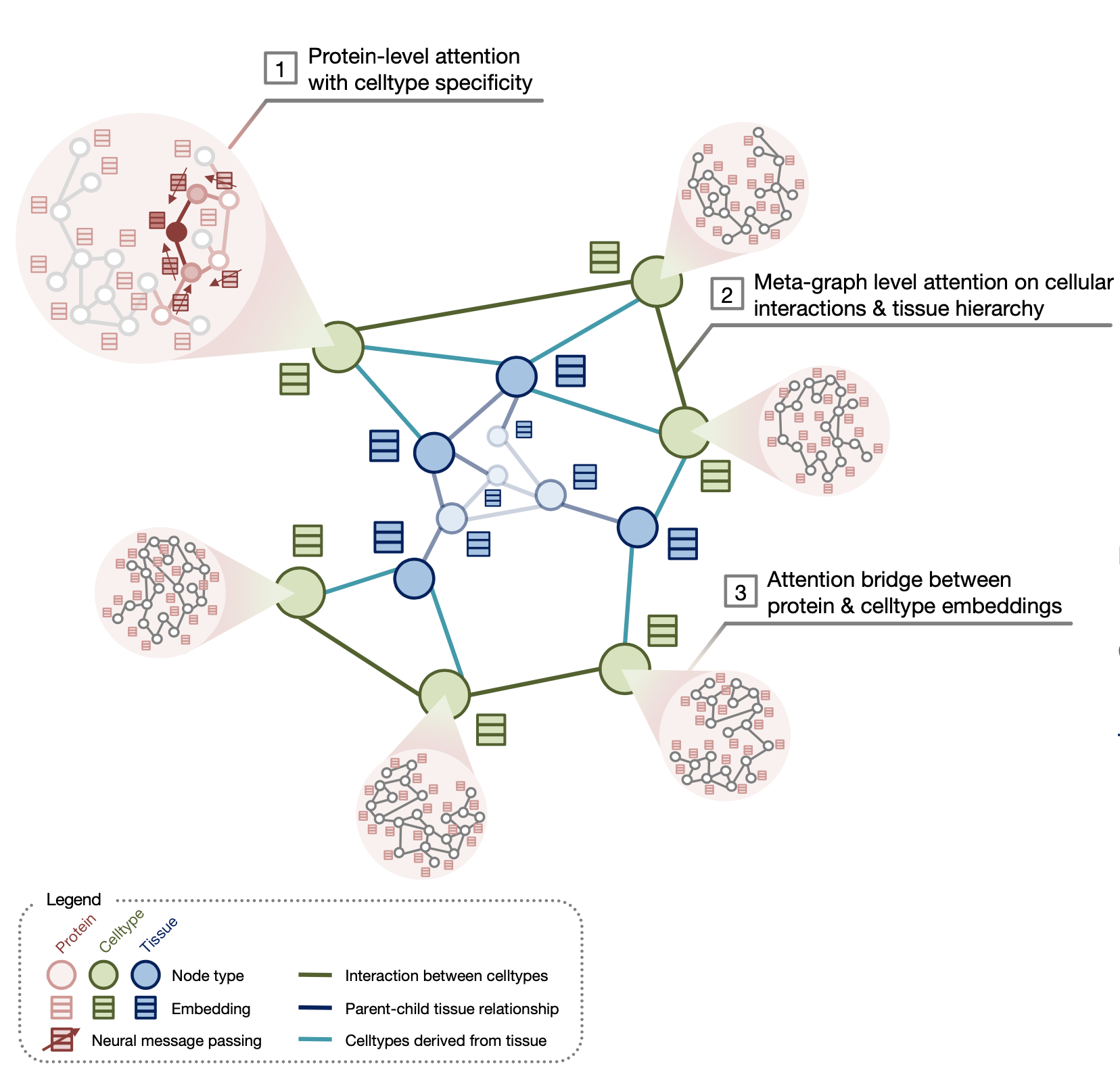
PINNACLE is a self-supervised geometric deep learning model that can generate protein representations in different celltype contexts. PINNACLE integrates single cell transcriptomics data with a protein interaction network, celltype interaction network, and tissue hierarchy to generate protein representations with celltype resolution.
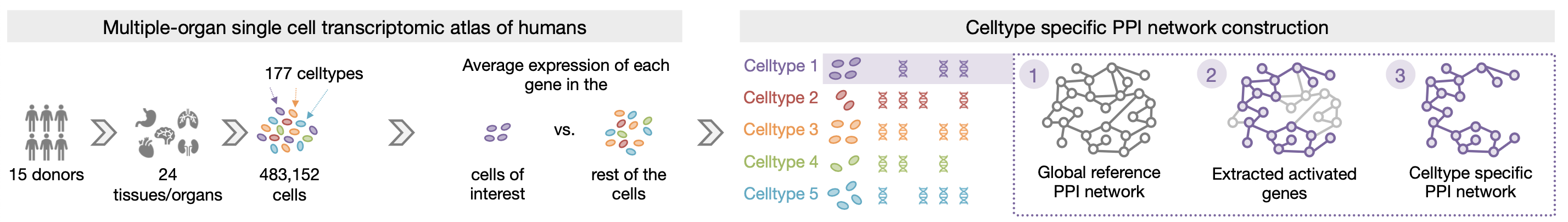
In this work, we focus on protein-coding genes and do not encode differences of protein isoforms (e.g., due to alternative splicing). Unlike existing methods that provide only one representation per protein (assuming each protein-coding gene encodes only one protein), resulting in fewer than 22,000 protein representations, PINNACLE generates a unique representation for each celltype that a protein is activated in.
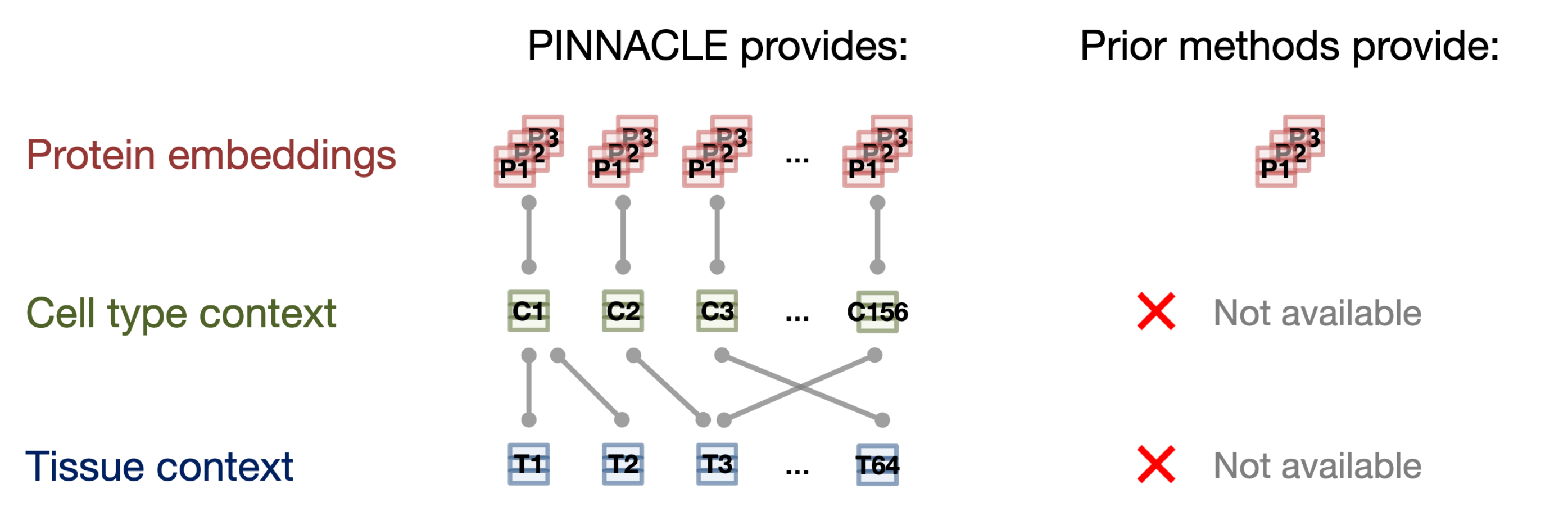
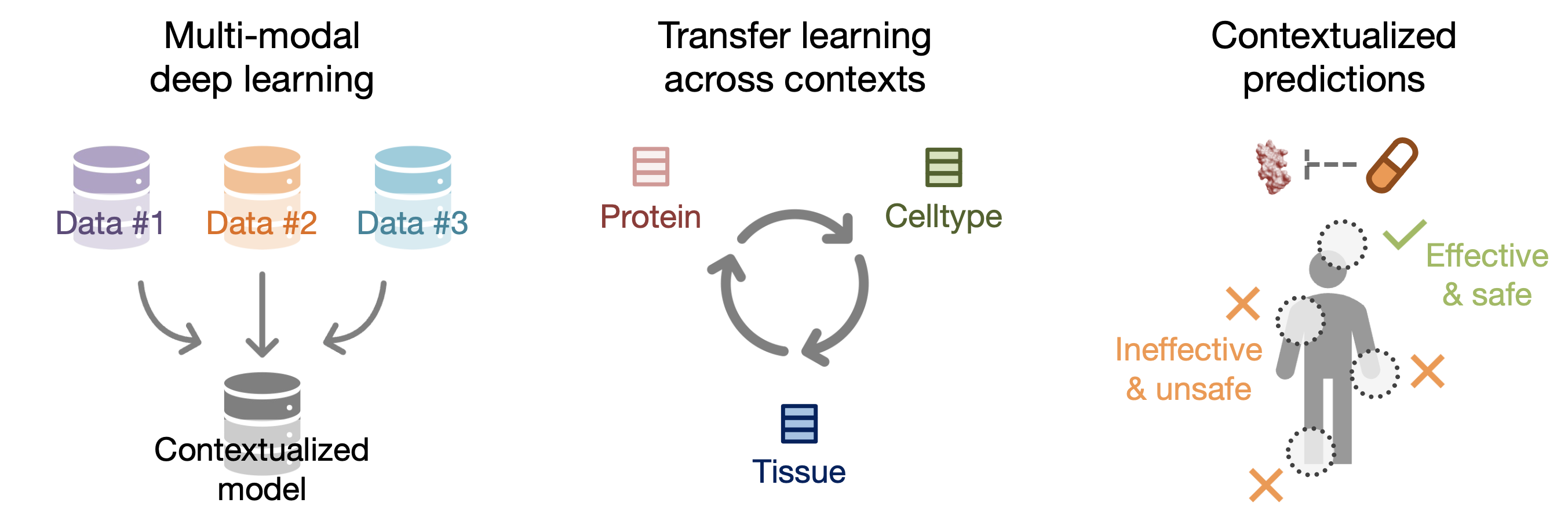
With our 394,760 contextualized protein representations (i.e., protein representations injected with celltype-specificity), we demonstrate PINNACLE’s ability to integrate structured and transcriptomic data, perform transfer learning across proteins, celltypes, and tissues, and generate contextualized predictions for diverse biomedical tasks.
PINNACLE Algorithm
PINNACLE is a self-supervised geometric deep learning model that can generate protein representations in diverse celltype contexts. PINNACLE is trained on a set of celltype specific protein interaction networks unified by a cellular and tissue network to produce contextualized protein representations based celltype activation. Unlike existing approaches, which do not consider biological context, PINNACLE produces multiple representations of proteins based on context, representations of the celltypes themselves, and representations of the tissues from which the celltypes are derived and the tissue hierarchy.
Given the multi-scale nature of the model inputs, PINNACLE is equipped to learn protein-level, celltype-level, and tissue-level topology in a single unified embedding space. To fully leverage the multi-scale inputs, PINNACLE uses protein-, celltype-, and tissue-level attention mechanisms and objective functions to inject cellular and tissue organization into the embedding space. PINNACLE is designed such that pairs of nodes that share an edge are embedded nearby each other, protein representations of the same celltype are embedded close by (and far from protein representations of a different celltype), and protein representations are embedded close to the representation of their corresponding celltype (and far from other celltype representations).
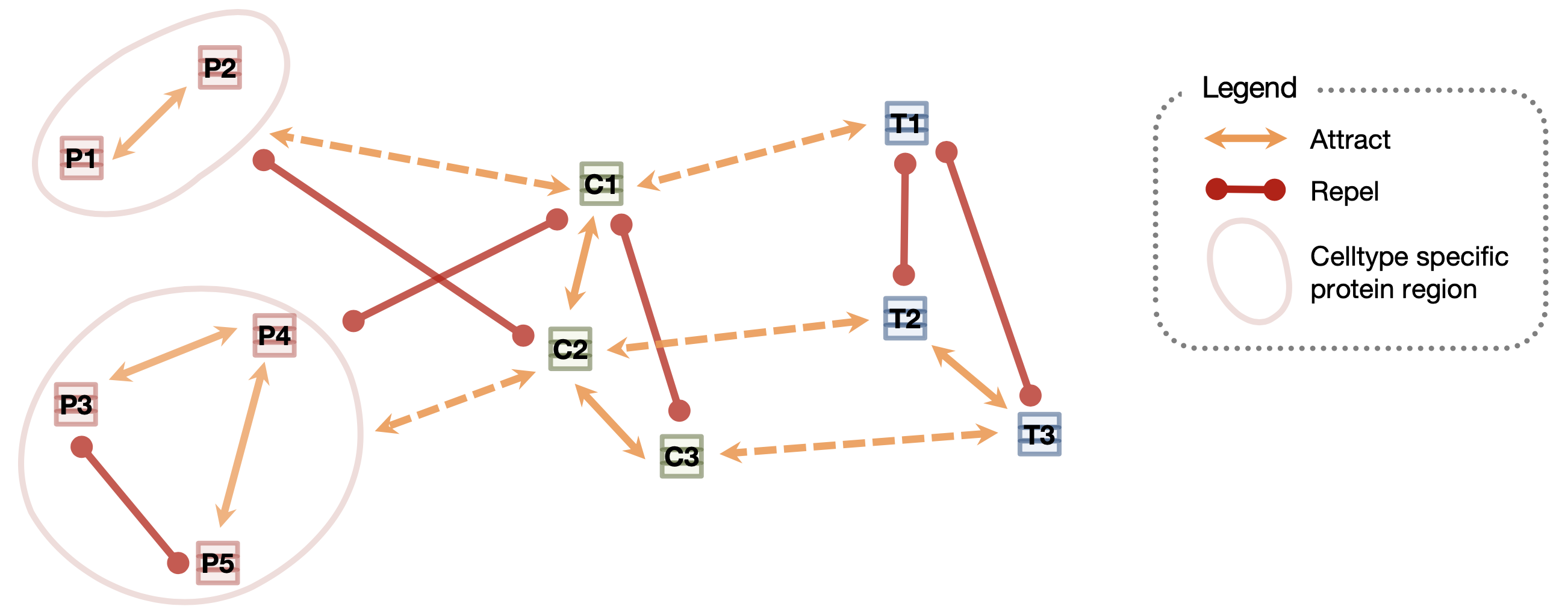
PINNACLE propagates message on proteins, celltypes, and tissues using attention mechanisms specific to each node and relationship type:
- The protein-level objective function, which considers self-supervised link prediction on the protein interactions and celltype-identity classification on the protein nodes, enables PINNACLE to produce an embedding space that captures both the topology of the celltype-specific protein interaction networks and the celltype identity of proteins.
- The celltype- and tissue-specific components in celltype- and tissue-specific objective functions are based solely on self-supervised link prediction to learn cellular and tissue organization.
- Such information is propagated to the protein representations using an attention bridge, imposing tissue and cellular organization to the protein representations.
Publication
Contextual AI Models for Single-Cell Protein Biology
Michelle M. Li, Yepeng Huang, Marissa Sumathipala, Man Qing Liang, Alberto Valdeolivas, Ashwin N. Ananthakrishnan, Katherine Liao, Daniel Marbach and Marinka Zitnik
Nature Methods 2024 [Paper] [bioRxiv]
@article{li2024contextualizing,
title={Contextual AI Models for Single-Cell Protein Biology},
author={Li, Michelle M and Huang, Yepeng and Sumathipala, Marissa and Liang, Man Qing and Valdeolivas, Alberto and Ananthakrishnan, Ashwin N and Marbach, Daniel and Zitnik, Marinka},
journal={Nature Methods},
url={https://www.nature.com/articles/s41592-024-02341-3},
year={2024}
}
Code Availability
Pytorch implementation of PINNACLE is available in the GitHub repository.
We provide an interactive demo to explore PINNACLE’s protein representations through a visual interface in the HuggingFace Space.