Biological networks are powerful representations for the discovery of molecular phenotypes. Fundamental to network analysis is the principle—rooted in social networks—that nodes that interact in the network tend to have similar properties. While this long-standing principle underlies powerful methods in biology that associate molecules with phenotypes on the basis of network proximity, interacting molecules are not necessarily similar, and molecules with similar properties do not necessarily interact.
Here, we show that molecules are more likely to have similar phenotypes, not if they directly interact in a molecular network, but if they interact with the same molecules. We call this the mutual interactor principle and show that it holds for several kinds of molecular networks, including protein-protein interaction, genetic interaction, and signaling networks.
We develop a machine learning framework for predicting molecular phenotypes on the basis of mutual interactors. Strikingly, the framework can predict drug targets, disease proteins, and protein functions in different species, and it performs better than much more complex algorithms. The framework is robust to incomplete biological data and is capable of generalizing to phenotypes it has not seen during training. Our work represents a network-based predictive platform for phenotypic characterization of biological molecules.
Molecules in and across living cells are constantly interacting, giving rise to complex biological networks. These networks serve as a powerful resource for the study of human disease, molecular function and drug-target interactions. For instance, evidence from multiple sources suggests that causative genes from the same or similar diseases tend to reside in the same neighborhood of protein-protein interaction networks. Similarly, proteins associated with the same molecular functions form highly-connected modules within protein-protein interaction networks.
These observations have motivated the development of bioinformatics methods that use molecular networks to infer associations between proteins and molecular phenotypes, including diseases, molecular functions, and drug targets. Many of these methods assume that molecular networks obey the organizing principle of homophily: the idea that similarity breeds connection. However, while this principle has been well-documented in social networks of many types (e.g. friendship, work, co-membership), it is unclear whether it captures the dynamics of biological networks. If not, existing bioinformatics methods that assume homophily may not realize the full potential of biological networks for scientific discovery.
Motivation for Mutual Interactors
To better understand the place for homophily in bioinformatics, we consider groups of phenotypically similar molecules (e.g. molecules associated with the same disease, involved in the same function, or targeted by the same drug) and study their interactions in large-scale biological networks. We find that most molecules associated with similar phenotypes do not interact directly in molecular networks, a result which puts into question the assumption of homophily, an assumption that is taken for granted by so many bioinformatics methods.
In fact, a different principle better explains how phenotypic similarity relates to network structure in biology. On average, two molecules that interact directly with one another will have less in common than two molecules that share many mutual interactors, just as people in a social network may share mutual friends. We call this the mutual interactor principle and validate it empirically on a diverse set of biological networks (see Figure 1c).
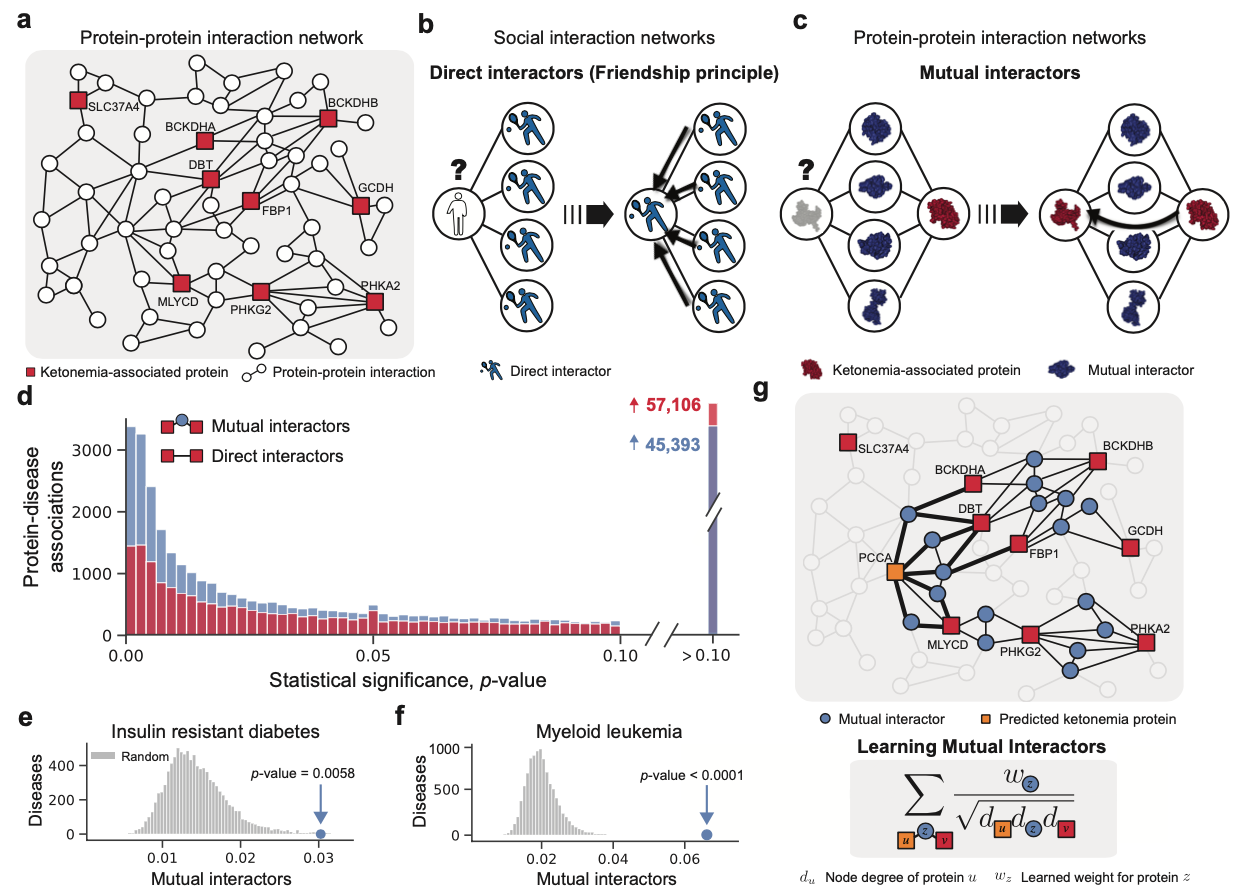
Mutual Interactors Approach
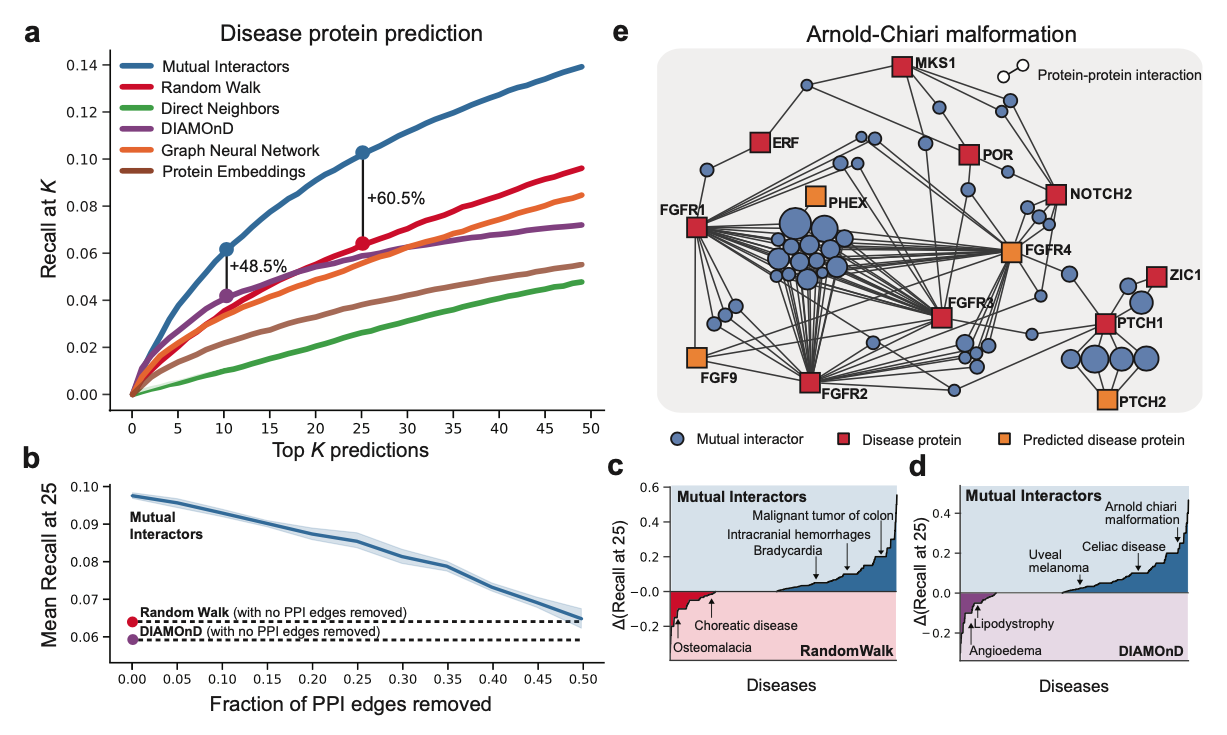
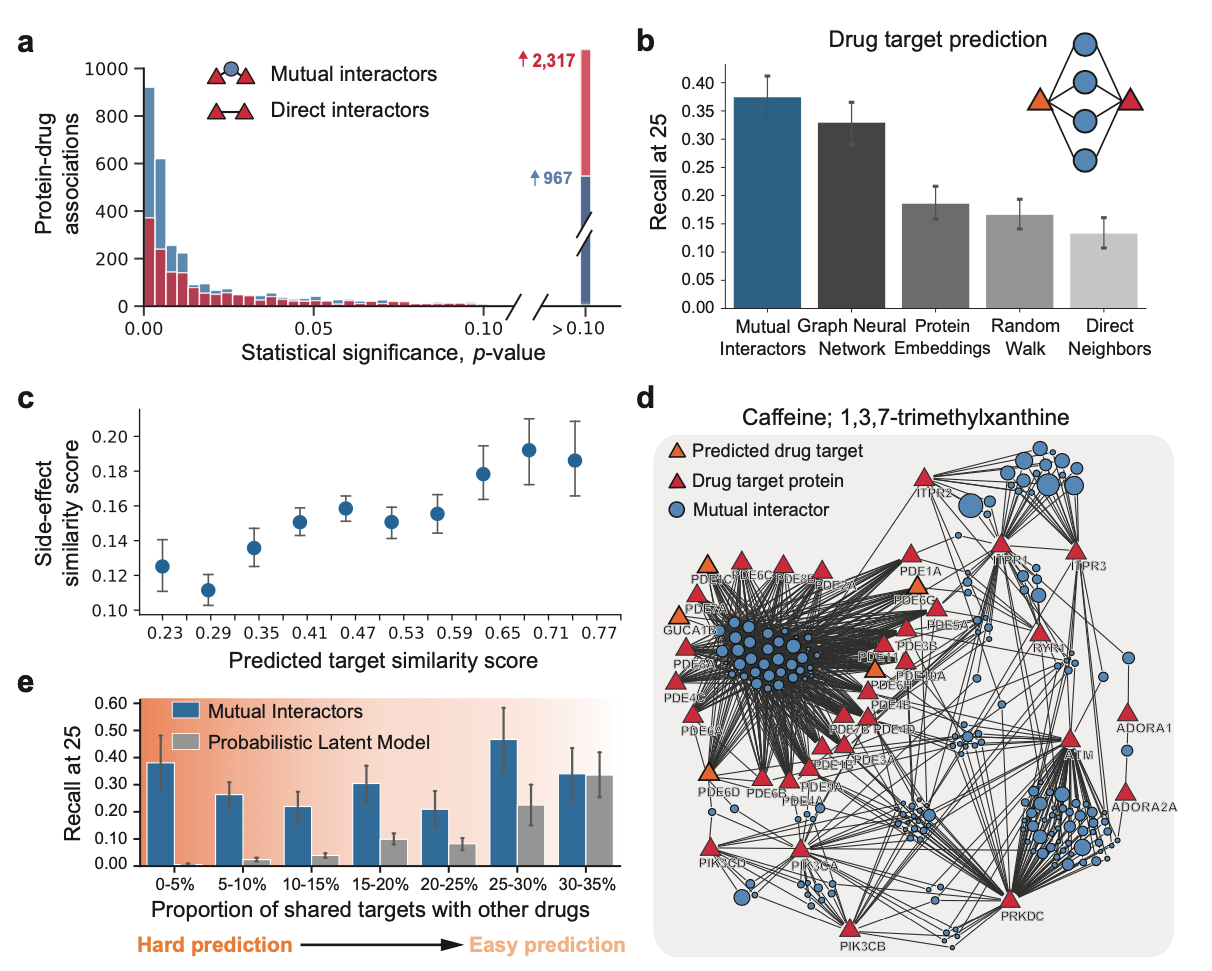
Motivated by our findings, we develop a machine learning framework, Mutual Interactors, that can predict a molecule’s phenotype based on the mutual interactors it shares with other molecules. We demonstrate the power, robustness, and scalability of Mutual Interactors on three key prediction tasks: disease protein prediction, drug target identification, and protein function prediction. With experiments across three different kinds of molecular networks (protein-protein interaction, signaling and genetic interaction) and four species (H. sapiens, S. cerevisiae, A. thaliana, M. musculus), we find that Mutual Interactors substantially outperforms existing methods, with gains in recall up to 61%. Additionally, we show that the weights learned by our method provide insight into the functional properties and druggability of mutual interactors.
Mutual Interactors is an approach based on a different network principle than existing bioinformatics methods. That it can outperform state-of-the-art approaches suggests a need to rethink the fundamental assumptions underlying machine learning methods for network biology
Uncovering Disease-Driving Proteins Using Mutual Interactors
Identifying Drug Targets Using Mutual Interactors
Publication
Mutual Interactors as a Principle for Phenotype Discovery in Molecular Interaction Networks
Sabri Eyuboglu*, Marinka Zitnik*, and Jure Leskovec
Pacific Symposium on Biocomputing, PSB 2023
@inproceedings{eyuboglu2023mutual,
title = {Mutual Interactors as a Principle for Phenotype Discovery in Molecular Interaction Networks},
author = {Eyuboglu, Sabri and Zitnik, Marinka and Leskovec, Jure},
booktitle = {Pacific Symposium on Biocomputing, PSB},
year = {2023}
}
Code
Python implementation of Mutual Interactors are available in the GitHub repository.