GraphXAI is a resource to systematically evaluate and benchmark the quality of GNN explanations. A key component is a novel and flexible synthetic dataset generator called ShapeGGen that can automatically generate a variety of benchmark datasets (e.g., varying graph sizes, degree distributions, homophilic vs. heterophilic graphs) together with ground-truth explanations that are robust to known pitfalls of explainable algorithms.
As graph AI models are increasingly used in high-stakes applications, it becomes essential to ensure that the relevant stakeholders can understand and trust their functionality. Only if the stakeholders clearly understand the behavior of these models, they can evaluate when and how much to rely on these models, and detect potential biases or errors in them. To this end, several approaches have been proposed to explain the predictions of GNNs. Based on the techniques they employ, these approaches can be broadly categorized into perturbation-based, gradient-based, and surrogate-based models.
To ensure that GNN explanations are reliable, it is important to correctly evaluate their quality. However, evaluating the quality of GNN explanations is a rather nascent research area with relatively little work. The approaches proposed thus far mainly leverage ground-truth explanations associated with specific datasets. However, this strategy is prone to several pitfalls:
-
For instance, there could be multiple underlying rationales (redundant/non-unique explanations) that could generate the true class labels and a given ground-truth explanation may only capture one of those, but the GNN model trained on the data may be relying on an entirely different rationale. In such a case, evaluating the explanation output by a state-of-the-art method using the ground-truth explanation is incorrect because the underlying GNN model itself is not relying on that ground-truth explanation.
-
In addition, even if there is a unique ground-truth explanation which generates the true class labels, the GNN model trained on the data could be a weak predictor which uses an entirely different rationale for making predictions. Post hoc explanations of such a model should not be evaluated based on the ground-truth explanation either.
-
Lastly, the ground-truth explanations corresponding to some of the existing benchmark datasets can be recovered using trivial baselines (e.g., random node or edge as explanation), and such datasets are not good candidates for reliably evaluating explanation quality.
Overview of GraphXAI
GraphXAI is a resource for systematic benchmarking and evaluation of GNN explainability methods. The process to evaluate explanation methods is to choose a graph problem and a GNN architecture to train, then train the GNN model and use a GNN explainer on its predictions to generate explanations. Finally, we compare explanations with a problem-given ground truth to provide a performance score for the GNN explainer. To this end, GraphXAI provides the following:
-
Dataset generator D} that can generate diverse types of graphs G, including homophilic, heterophilic, and attributed graphs suitable for the study of graph explainability. Prevailing benchmark datasets are designed for benchmarking GNN predictors and typically consist of a graph or a set of graphs and associated ground-truth label information. While these datasets are sufficient for studying GNN predictors, they cannot be readily used for studying GNN explainers because they lack a critical component, namely information on ground-truth explanations. GraphXAI addresses this critical gap by providing the SHapeGraph generator to create graphs with ground-truth explanations that are uniquely suited for studying GNN explainers.
-
GNN predictor f that is a user-specified GNN model trained on a dataset produced by D and optimized to predict labels for a particular downstream task.
-
GNN explanation method(s) O that takes a prediction f(u) and returns an explanation M(u) = O(f, u) for it.
-
Explanation quality metrics P such that each metric takes a set of explanations and evaluates them for correctness relative to ground-truth explanations.
When taken together, GraphXAI provides all the necessary functionality needed to systematically benchmark and evaluate GNN explainability methods. Further, it addresses the above mentioned pitfalls of state-of-the-art evaluation setups for GNN explanation methods.
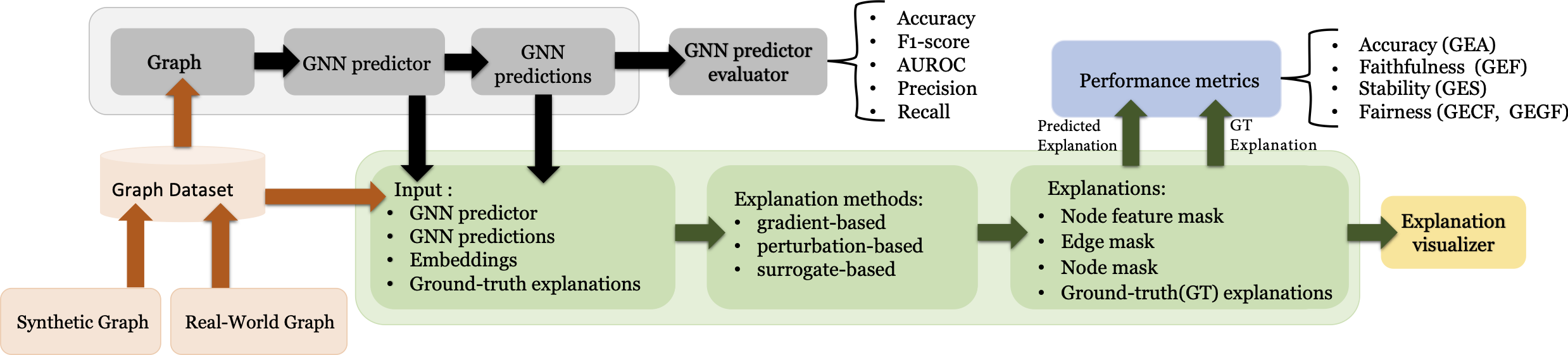
GraphXAI includes the following:
-
novel generator ShapeGGen to automatically generate diverse types of XAI-ready benchmark datasets, including homophilic, heterophilic, and attributed graphs, each accompanied by ground-truth explanations,
-
graph and explanation functions compatible with deep learning frameworks, such as PyTorch and PyTorch Geometric libraries,
-
training and visualization functions for GNN explainers,
-
utility functions to support the development of new GNN explainers, and
-
comprehensive set of performance metrics to evaluate the correctness of explanations produced by GNN explainers relative to ground-truth explanations.
ShapeGGen Data Generator
ShapeGGen is a generator of XAI-ready graph datasets supported by graph theory and particularly suitable for benchmarking GNN explainers and study their limitations.
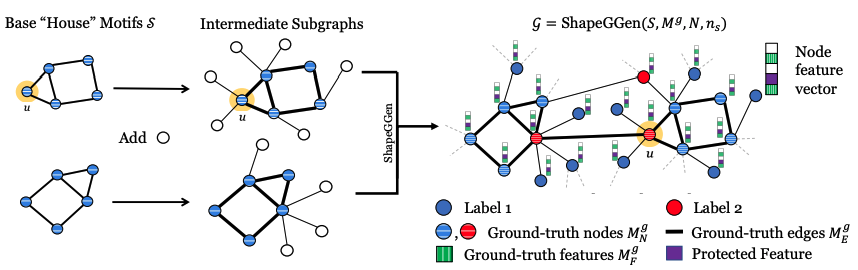
ShapeGGen generates graphs by combining subgraphs containing any given motif and additional nodes. The number of motifs in a k-hop neighborhood determines the node label (in the figure, we use a 1-hop neighborhood for labeling, and nodes with two motifs in its 1-hop neighborhood are highlighted in red). Feature explanations are some mask over important node features (green striped), with an option to add a protected feature (shown in purple) whose correlation to node labels is controllable. Node explanations are nodes contained in the motifs (horizontal striped nodes) and edge explanations (bold lines) are edges connecting nodes within motifs.
Publication
Evaluating Explainability for Graph Neural Networks
Chirag Agarwal*, Owen Queen*, Himabindu Lakkaraju and Marinka Zitnik
Scientific Data 2023 [arXiv]
* Equal Contribution
@article{agarwal2023evaluating,
title={Evaluating Explainability for Graph Neural Networks},
author={Agarwal, Chirag and Queen, Owen and Lakkaraju, Himabindu and Zitnik, Marinka},
journal={Scientific Data},
volume={10},
number={144},
url={https://www.nature.com/articles/s41597-023-01974-x},
year={2023},
publisher={Nature Publishing Group}
}
Code
Datasets and Pytorch implementation of GraphXAI are available in the GitHub repository.