Deep learning methods for graphs achieve remarkable performance on many tasks. However, despite the proliferation of such methods and their success, recent findings indicate that even the strongest and most popular Graph Neural Networks (GNNs) are highly vulnerable to adversarial attacks. Adversarial attacks mean that an attacker injects small but carefully-designed perturbations to the graph structures in order to degrade the performance of GNN classifiers.
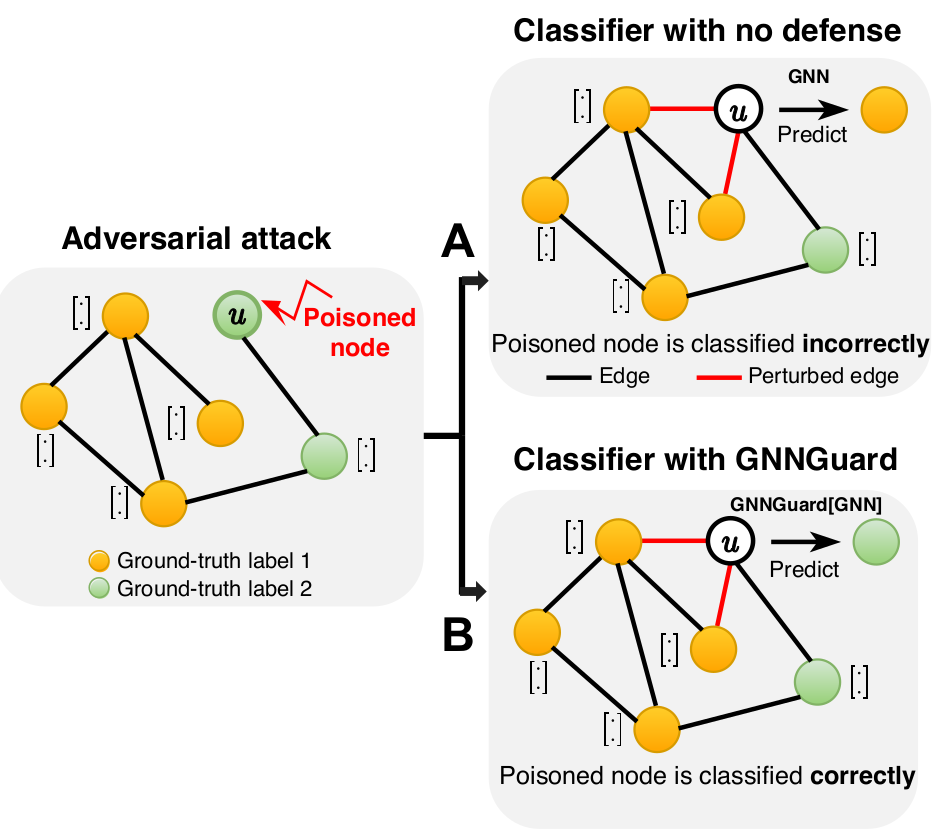
The vulnerability is a significant issue preventing GNNs from being used in real-world applications. For example, under adversarial attack, small and unnoticeable perturbations of graph structure (e.g., adding two edges on the poisoned node) can catastrophically reduce performance (panel A in the figure).
We develop GNNGuard, a general algorithm to defend against a variety of training-time attacks that perturb the discrete graph structure. GNNGuard can be straightforwardly incorporated into any GNN. By integrating GNNGuard, the GNN classifier can make correct predictions even when trained on the attacked graph (panel B in the figure).
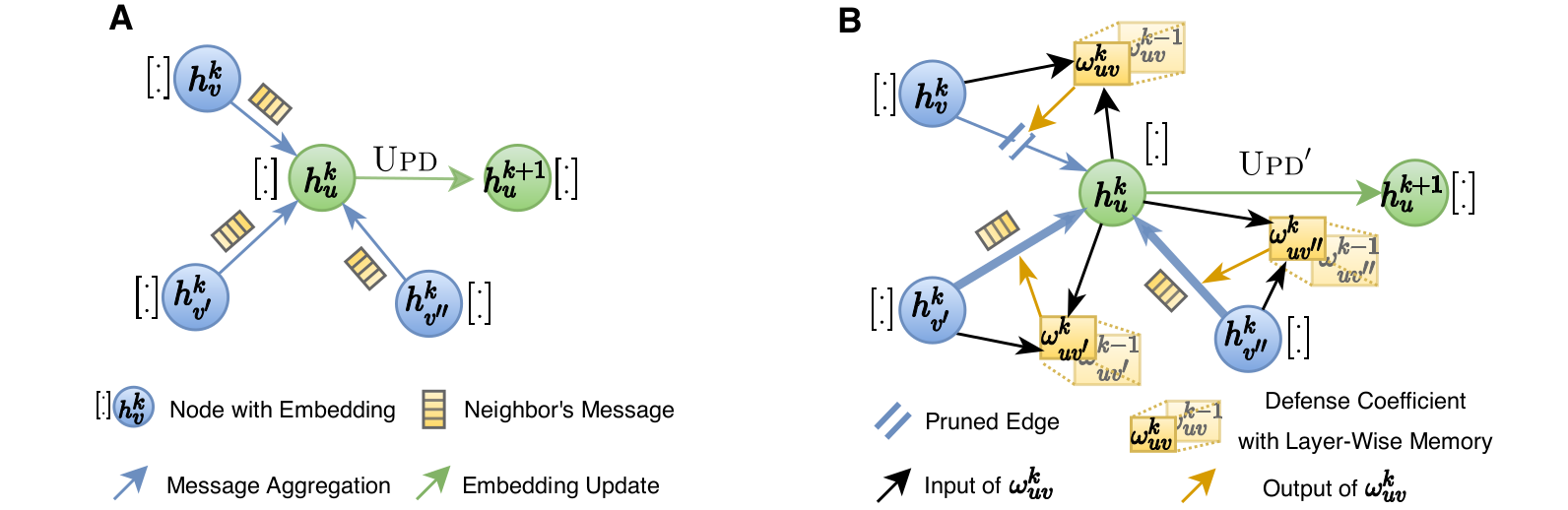
GNNGuard algorithm
Most damaging attacks add fake edges between nodes that have different features and labels. Because of that, the key idea of GNNGuard is to detect and quantify the relationship between the graph structure and node features, if one exists, and then exploit that relationship to mitigate negative effects of the attack. GNNGuard learns how to best assign higher weights to edges connecting similar nodes while pruning edges between unrelated nodes. In specific, instead of the neural message passing of a typical GNN (panel A in the figure), GNNGuard (panel B in the figure) controls the message stream, such as blocking the message from irrelevant neighbors while strengthening messages from highly-related ones.
Remarkably, GNNGuard can effectively restore state-of-the-art performance of GNNs in the face of various adversarial attacks, including targeted and non-targeted attacks, and can defend against attacks on both homophily and heterophily graphs.
Attractive properties of GNNGuard
- Defense against a variety of attacks: GNNGuard is a general defense approach that is effective against a variety of training-time attacks, including directly targeted, influence, and non-targeted attacks.
- Integrates with any GNNs: GNNGuard can defend any modern GNN architecture against adversarial attacks.
- State-of-the-art performance on clean graphs: In real-world settings, we do not know whether a graph has been attacked or not. GNNGuard can restore state-of-the-art performance of a GNN when the graph is attached as well as sustain the original performance on non-attacked graphs.
- Homophily and heterophily graphs: GNNGuard is the first technique that can defend GNNs against attacks on homophily and heterophily graphs. GNNGuard can be easily generalized to graphs with abundant structural equivalences, where connected nodes have different node features yet similar structural roles.
Publication
GNNGuard: Defending Graph Neural Networks against Adversarial Attacks
Xiang Zhang and Marinka Zitnik
NeurIPS 2020 [arXiv] [poster]
@inproceedings{zhang2020gnnguard,
title = {GNNGuard: Defending Graph Neural Networks against Adversarial Attacks},
author = {Zhang, Xiang and Zitnik, Marinka},
booktitle = {Proceedings of Neural Information Processing Systems, NeurIPS},
year = {2020}
}
Code and datasets
Pytorch implementation of GNNGuard and all datasets are available in the GitHub repository.