Integrating structured clinical knowledge into artificial intelligence (AI) models remains a major challenge. Medical codes primarily reflect administrative workflows rather than clinical reasoning, limiting AI models’ ability to capture true clinical relationships and undermining their generalizability.
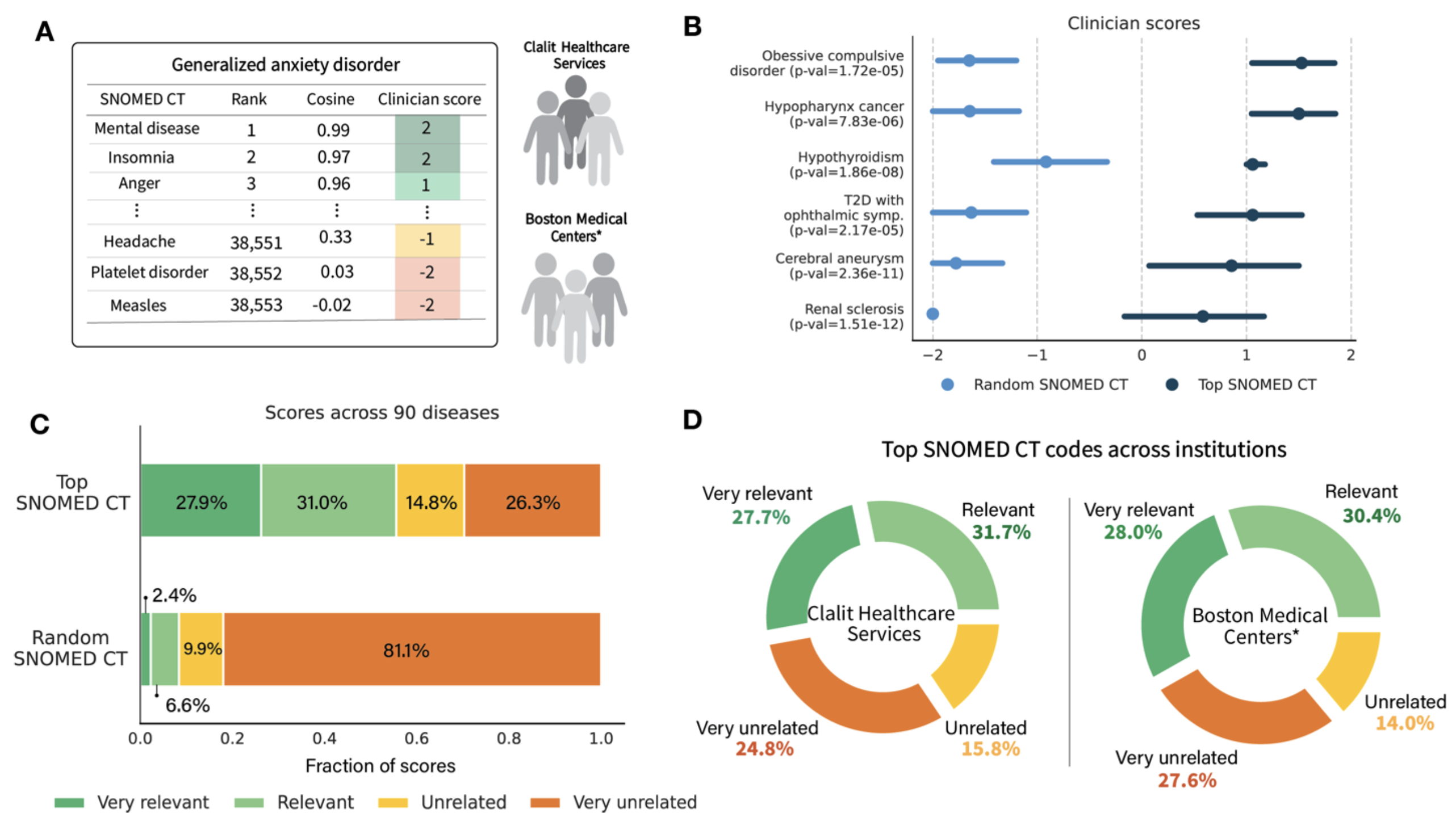
To address this, we introduce ClinGraph, a clinical knowledge graph that integrates eight EHR-based vocabularies, and ClinVec, a set of 153,166 clinical code embeddings derived from ClinGraph using a graph transformer neural network. ClinVec provides a machine-readable representation of clinical knowledge that captures semantic relationships among diagnoses, medications, laboratory tests, and procedures. Panels of clinicians from multiple institutions evaluated the embeddings across 96 diseases and more than 3,000 clinical codes, confirming their alignment with expert knowledge.
In a retrospective analysis of 4.57 million patients from Clalit Health Services, we show that ClinVec supports phenotype risk scoring and stratifies individuals by survival outcomes. We further demonstrate that injecting ClinVec into large language models improves performance on medical question answering, including for region-specific clinical scenarios. ClinVec enables structured clinical knowledge to be injected into predictive and generative AI models, bridging the gap between EHR codes and clinical reasoning
Introduction
Medicine is grounded in centuries of medical knowledge and the pursuit of individualized patient care through meticulous reasoning and evidence-based practice. The extensive development of standardized vocabularies and ontologies in the medical field has fostered a unified representation of clinical information, enabling interoperability and data exchange across healthcare systems globally. These standardized coding systems provide a consistent framework for effectively representing clinical knowledge, forming the backbone of precision medicine initiatives. With the widespread adoption of electronic health records (EHRs) and the standardization of medical data, precision medicine has shifted toward large-scale, data-driven approaches that directly leverage structured EHR data. More than half of healthcare foundation models now rely exclusively on structured clinical codes, such as billing data and medication records.
we developed a ClinGraph, a clinical knowledge graph spanning eight standardized medical vocabularies, and ClinVec, a set of 153,166 clinical code embeddings derived from ClinGraph. Using state-of-the-art relational graph transformers and a clinical knowledge graph, we created a cohesive, machine-readable map that captures relationships among seven clinical vocabularies, including laboratory tests, diagnosis codes, and medications, without requiring manual curation. By integrating verified knowledge bases and medical ontologies into a knowledge graph of standardized EHR codes, this resource reduces the risk of propagating inaccuracies while ensuring interpretability and transparency.
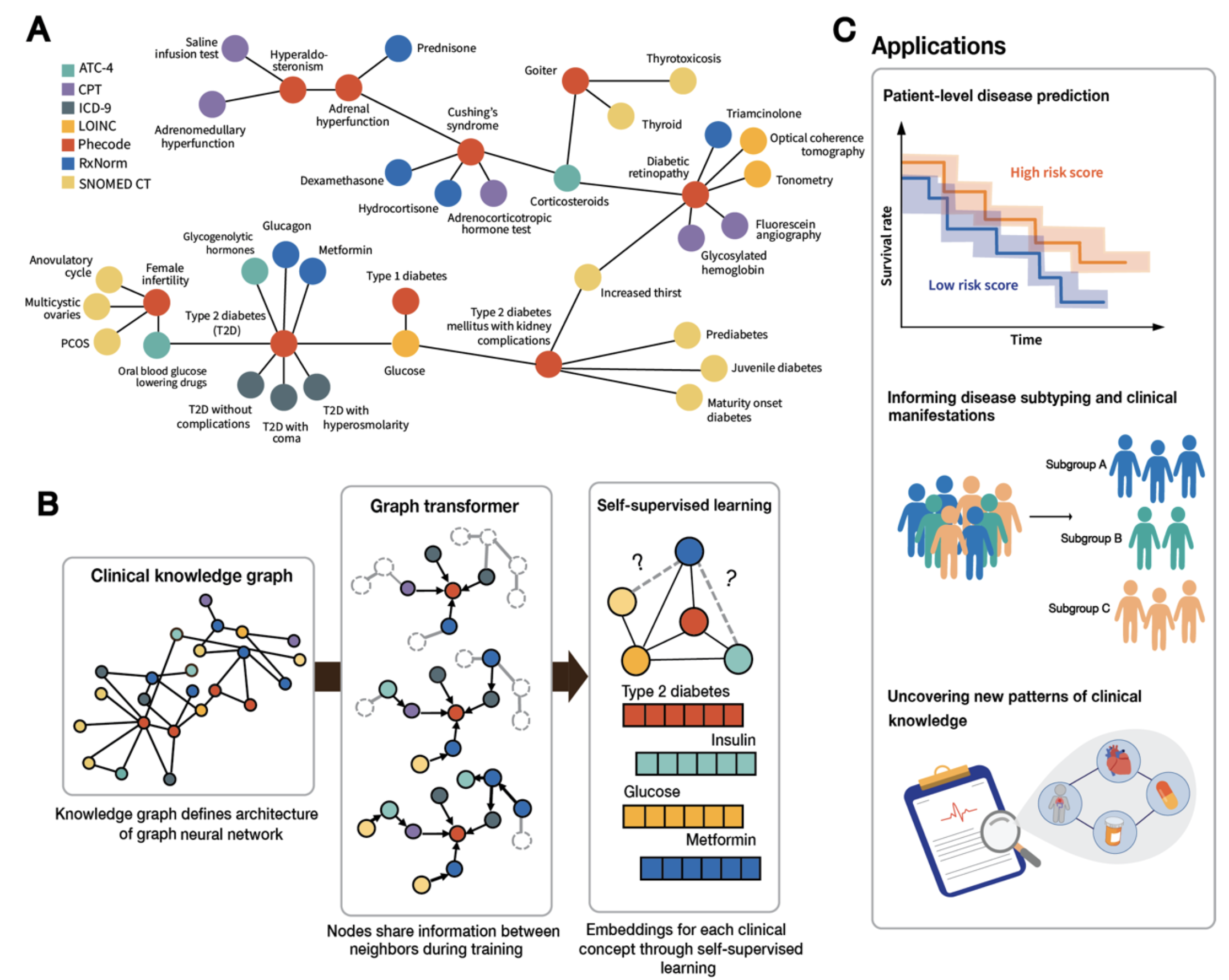
Our resource provides a hypothesis-free approach to generating clinically insightful representations of medical codes. It offers three main applications:
- Integrating clinical knowledge into precision medicine patient models,
- Enabling patient-agnostic generalizable models of populations that can be safely exchanged across institutions, and
- Providing insights into the organization of clinical knowledge.
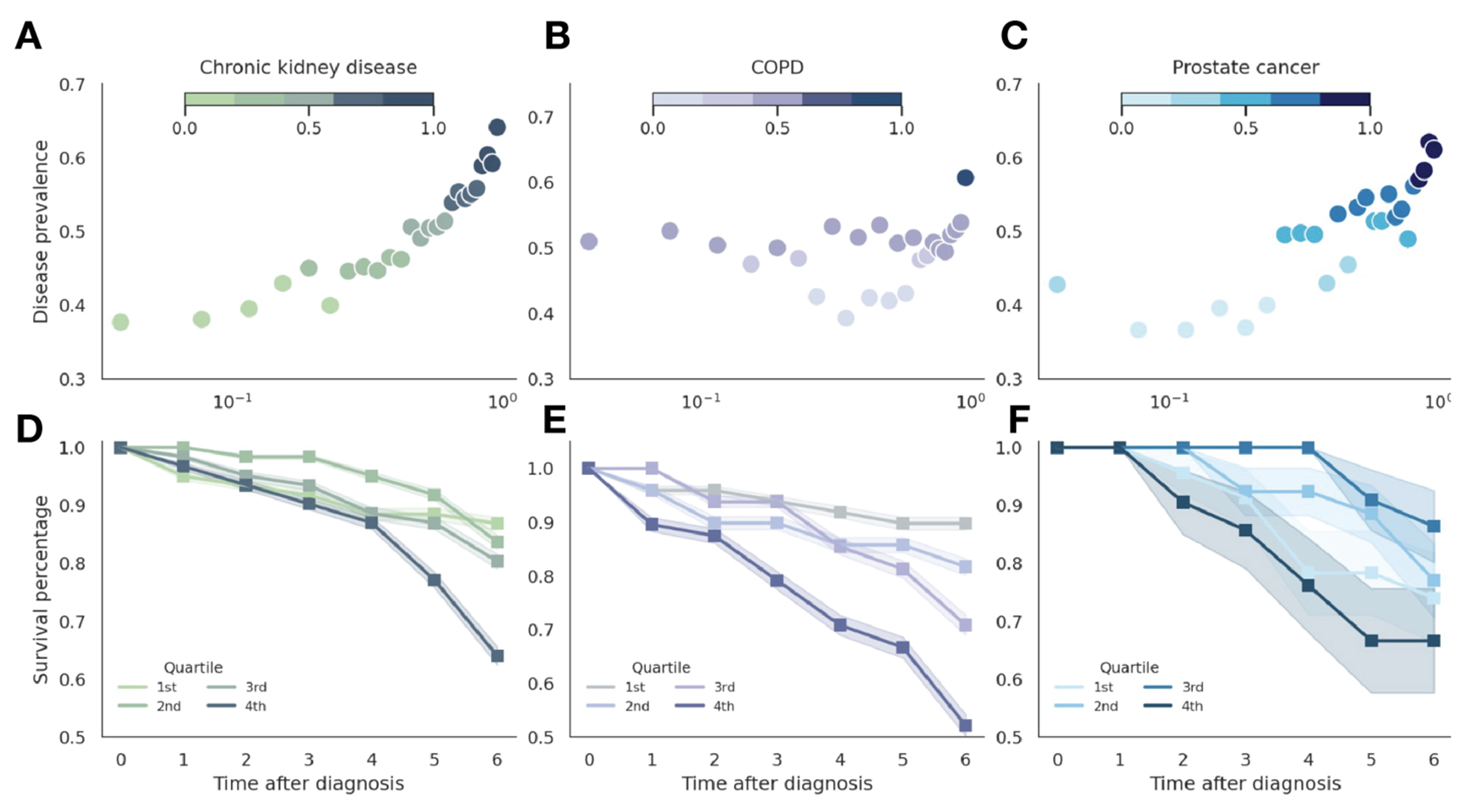
The latent space of medical codes reveals patterns consistent with human anatomy and disease presentations, capturing symptomatic and clinical presentations of diseases that can be decomposed into symptom embeddings. We demonstrate the predictive utility of these embeddings through a large-scale phenotype risk score analysis for three chronic diseases across 4.57 million patients from Clalit Healthcare Services.
An expert clinical evaluation across 90 diseases and 3,000 clinical codes conducted with clinician panels in the United States and Israel validates the alignment of these embeddings with established medical knowledge. Our findings establish unified medical code embeddings as a foundational resource for advancing AI-driven healthcare.
Unified clinical vocabulary embeddings can facilitate collaborative, scalable efforts in clinical AI and offer a tool for deepening our understanding of disease relationships, laboratory tests, diagnosis codes, medications, and their underlying mechanisms.
Clinical embeddings capture knowledge of human anatomy and clinical subspecialties
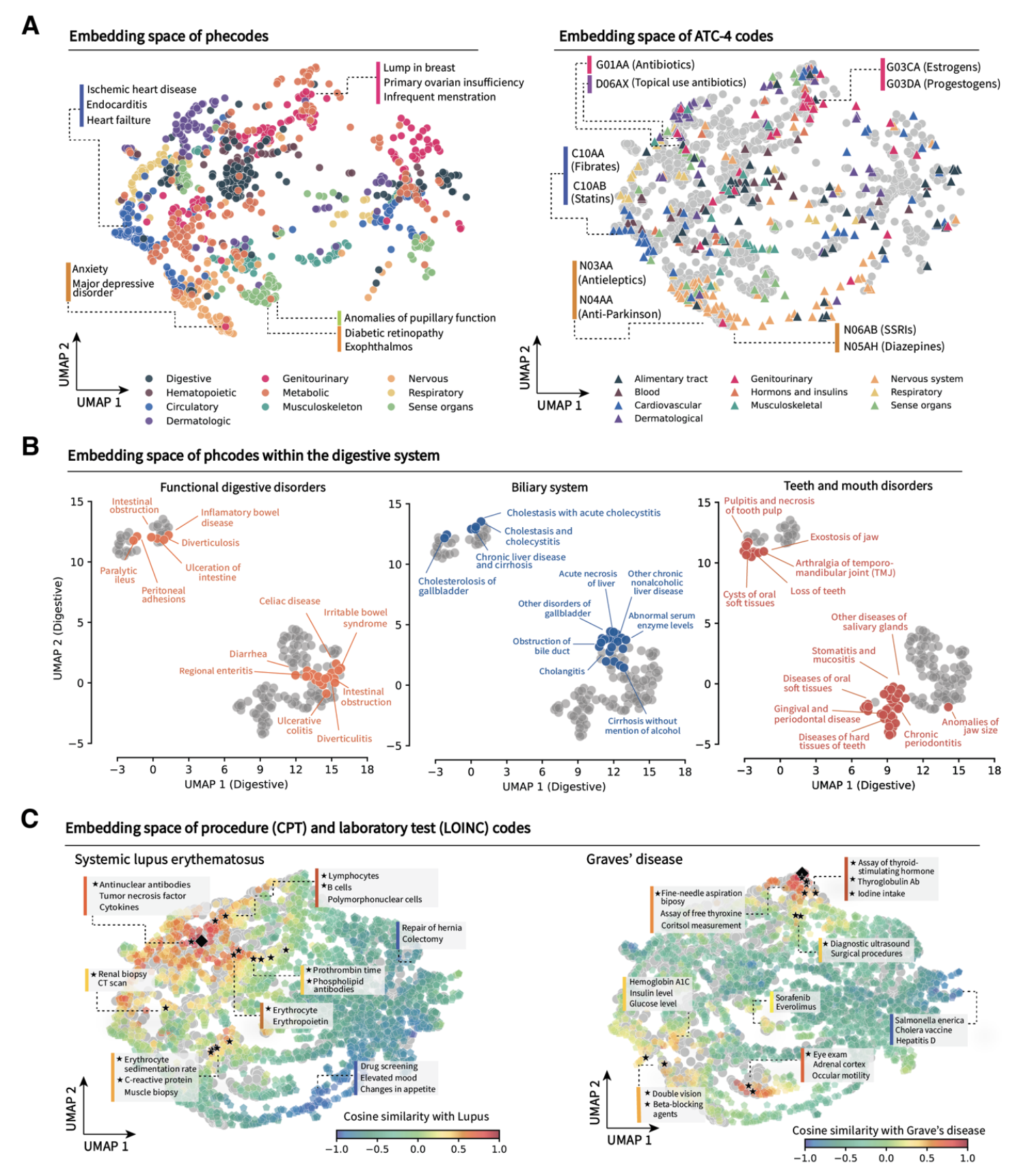
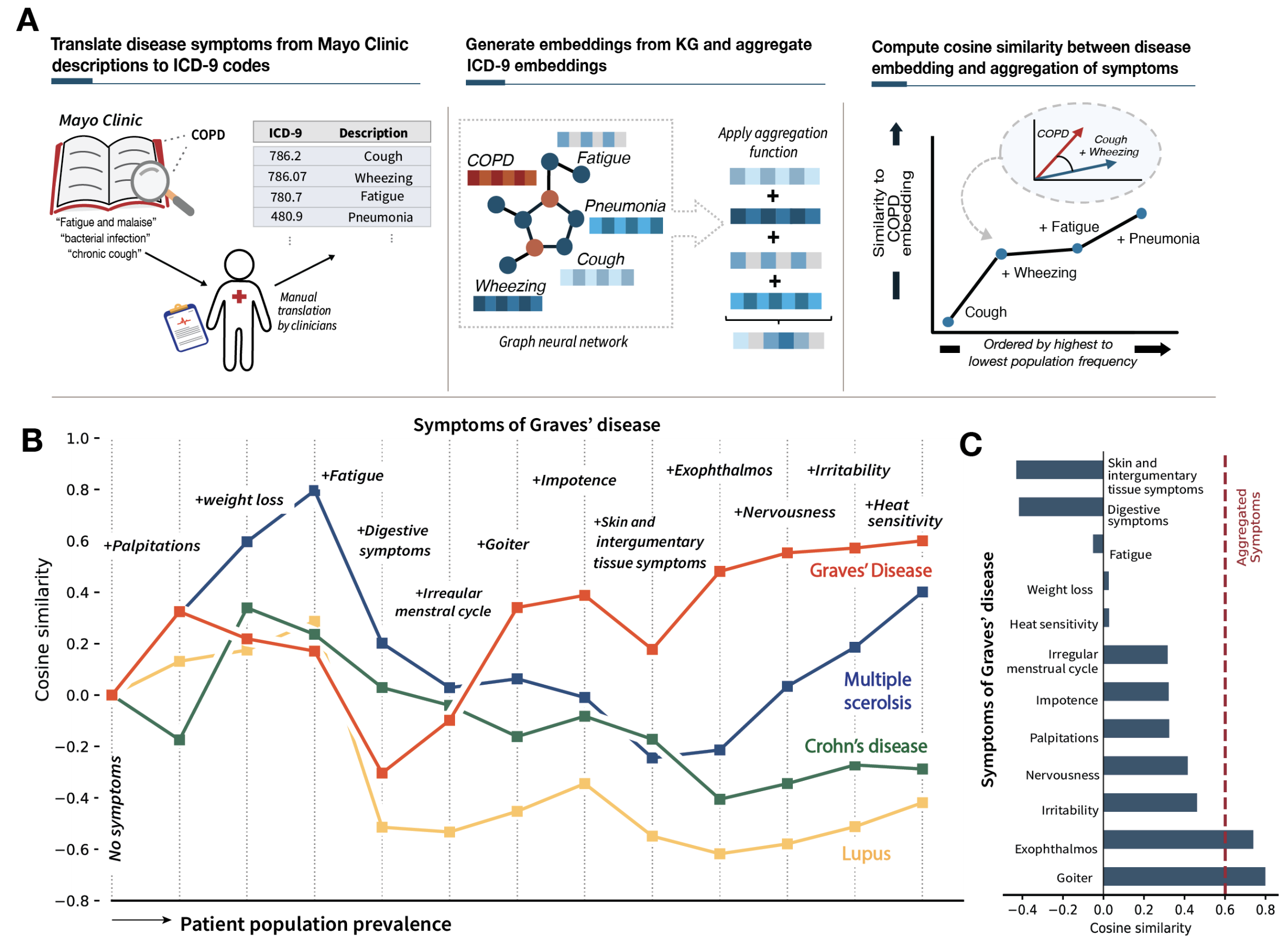
Latent embedding space reflects patterns of disease presentation and diagnostic processes
Unified vocabulary embeddings enable disease risk stratification and severity prediction
Clinical vocabulary embeddings capture medical knowledge consensus across institutions
Publication
ClinVec: Unified Embeddings of Clinical Codes Enable Knowledge-Grounded AI in Medicine
Ruth Johnson, Uri Gottlieb, Galit Shaham, Lihi Eisen, Jacob Waxman, Stav Devons-Sberro, Curtis R. Ginder, Peter Hong, Raheel Sayeed, Xiaorui Su, Ben Y. Reis, Ran D. Balicer, Noa Dagan, and Marinka Zitnik
In Review 2025 [medRxiv]
@article{johnson2024unified,
title={ClinVec: Unified Embeddings of Clinical Codes Enable Knowledge-Grounded AI in Medicine},
author={Johnson, Ruth and Gottlieb, Uri and Shaham, Galit and Eisen, Lihi and Waxman, Jacob and Devons-Sberro, Stav and Ginder, Curtis R. and Hong, Peter and Sayeed, Raheel and Reis, Ben Y. and Balicer, Ran D. and Dagan, Noa and Zitnik, Marinka},
journal={medrxiv},
url={https://www.medrxiv.org/content/10.1101/2024.12.03.24318322},
year={2024}
}
Code and Data Availability
Pytorch implementation of ClinVec and ClinGraph is available in the GitHub repository.
ClinVec embeddings and ClinGraph knowledge graph are available via Harvard Dataverse. Due to national and organizational data privacy regulations, CHS individual-level data from this study cannot be shared publicly.
Authors
- Ruth Johnson
- Uri Gottlieb
- Galit Shaham
- Lihi Eisen
- Jacob Waxman
- Stav Devons-Sberro
- Curtis R. Ginder
- Peter Hong
- Raheel Sayeed
- Ben Y. Reis
- Ran D. Balicer
- Noa Dagan
- Marinka Zitnik
This research would not be possible without the generous support by The Ivan and Francesca Berkowitz Family Living Laboratory Collaboration at Harvard Medical School and Clalit Research Institute.