Teaching
AIM 2: Artificial Intelligence in Medicine 2
AI continues to transform medicine, offering cutting-edge approaches to address challenges in medical research and practice. This course covers modern paradigms in AI, including self-supervised learning, generative models, and multimodal techniques with applications to natural language processing, medical image analysis, relational and structure understanding, and longitudinal sensor data.
Our study on the use of generative AI in education with prospective evaluation of knowledge tracing in the classroom.
BMI 702: Biomedical Artificial Intelligence
AI is poised to enable breakthroughs in science and reshape medicine. This introductory course provides a survey of artificial intelligence for biomedical informatics, covering methods for key data modalities: clinical data, networks, language, and images. It introduces machine learning problems from a practical perspective, focusing on tasks that drive the adoption of machine learning in biology and medicine.
It outlines foundational algorithms and emphasizes the subtleties of working with biomedical data and ways to evaluate and transition machine learning methods into biomedical and clinical implementation. An important consideration in this course is the broader impact of artificial intelligence, particularly topics of bias and fairness, interpretability, and ethical and legal considerations when dealing with artificial intelligence.
International Competitions
CUREBench (NeurIPS 2025)
Benchmarking AI reasoning for therapeutic decision-making at scale. This competition is colocated with
the International Conference on Neural Information Processing Systems (NeurIPS).
International Workshops and Conferences
AI Virtual Cells and Instruments: A New Era in Drug Discovery and Development (NeurIPS 2025)
This workshop focuses on building virtual cells and instruments with AI to support large-scale simulation and analysis of molecules, cells, and tissues. We define this emerging approach and examine how it can be applied to improve drug discovery and development.
This workshop is presented at the International Conference on Neural Information Processing Systems (NeurIPS).
AI for Science: The Reach and Limits of AI for Scientific Discovery (NeurIPS 2025)
This workshop maps the current capabilities of AI models across scientific domains and explores how to push them further. We focus on three central challenges. First, we ask whether today’s large language models generate rigorously testable hypotheses and reason over experimental results spanning fields like physics, chemistry, and biology. Second, we examine the fidelity of generative and surrogate simulators. In biology, all-atom models gain power; in chemistry, machine learning force fields improve in accuracy and generalizability; and in climate science, models predict weather up to 15 days ahead. We consider how far these advances reach and which spatial or temporal scales remain intractable. Third, we address experimental data scarcity and bias. Consortium-driven efforts like the Protein Data Bank, Human Cell Atlas, Materials Project, and Open Molecules Dataset drive progress, but many fields still lack large-scale datasets. We ask where AI can benefit most from new data generation, how far models trained on limited data can take us, and when lab-in-the-loop strategies are essential.
This workshop is presented at the International Conference on Neural Information Processing Systems (NeurIPS).
Harvard AI for Genomics and Health (18th Annual PQG Conference)
Recent breakthroughs at the nexus of AI, genomics, and healthcare have been remarkable. Cutting-edge technologies, including transformers, large language models, and protein structure models, are leveraging vast datasets to exhibit remarkable emergent capabilities. At the same time, biotechnological innovations such as high-throughput CRISPR screens, multiplex imaging, and spatial omics are producing rich data at unprecedented scales and resolutions.
AI for Science: Scaling in AI for Scientific Discovery (ICML 2024)
As AI extends its reach to a broader range of scientific questions, we explore the role of scalable AI in current scientific endeavors: what further contributions can we expect from AI in research? How can AI techniques be effectively harnessed? Moreover, how does AI influence the objectives and methods of science?
This workshop is presented at the International Conference on Machine Learning (ICML).
AI for Scientific Discovery: From Theory to Practice (NeurIPS 2023)
For centuries, the method of discovery—the fundamental practice of science that scientists use to systematically and logically explain the natural world—has remained largely unchanged. Artificial intelligence and machine learning hold tremendous promise for revolutionizing the way scientific discovery is conducted at a fundamental level. However, to realize this potential, it's imperative to identify priorities and address the pressing open questions that stand at the cutting edge of AI. We are especially keen on exploring topics such as solving grand challenges in structural biology, scaling dynamical system modeling to millions of particles, visualizing the previously unimaginable black hole, incorporating physical insights into AI methods, and accelerating drug discovery.
This workshop is presented at the International Conference on Neural Information Processing Systems (NeurIPS).
Graph Learning Benchmarks (KDD 2023)
The meeting aims to address the limitations of current benchmark datasets in graph machine learning and the bias towards certain directions in the development of graph learning techniques. To increase the diversity of graph learning benchmarks, identify new research directions, and better understand how different techniques perform on these benchmarks, the meeting discusses novel tasks and datasets. Unlike most existing graph learning workshops that focus on developing new models and algorithms, the meeting focuss on specifying new datasets, tasks, and applications and formalizing them as new benchmarks.
The workshop is presented at the KDD conference.
Graph Representations and Algorithms in Biomedicine (PSB 2023)
Connectivity is a fundamental property of biological systems: on the cellular level, proteins interact with each other to form protein-protein interaction networks; on the organism level, neurons are arranged in a network; and on a community level, species can have complex relationships with one another that drive the development of an ecosystem. Graphs, mathematical representations modeling entities as vertices and their relationships as edges, have proved useful for understanding biological systems that naturally have such a network structure. Graph representations and algorithms (often in combination with machine learning techniques) can be used to organize massive amounts of related (and sometimes heterogenous or unstructured) data, and to ultimately to identify patterns that represent novel biological insights.
This session will encompass modern developments in graph theory, computational topology, and graph machine learning applied to various fields of biomedicine. This workshop is presented at the Pacific Symposium on Biocomputing (PSB).
AI4Science: Progress and Promises (NeurIPS 2022)
For centuries, the method of discovery—the fundamental practice of science that scientists use to explain the natural world systematically and logically—has remained largely the same. Artificial intelligence (AI) and machine learning (ML) hold tremendous promise in having an impact on the way scientific discovery is performed today at the fundamental level. However, to realize this promise, we need to identify priorities and outstanding open questions for the cutting edge of AI going forward.
This workshop is presented at the International Conference on Neural Information Processing Systems (NeurIPS).
New Frontiers in Graph Learning (NeurIPS 2022)
Despite the success, existing graph learning paradigms have not captured the full spectrum of relationships in the physical and the virtual worlds. For example, in terms of applicability of graph learning algorithms, current graph learning paradigms are often restricted to datasets with explicit graph representations, whereas recent works have shown promise of graph learning methods for applications without explicit graph representations. Further, in terms of generalizability, unlike domains such as computer vision and natural language processing where large-scale pre-trained models generalize across downstream applications with little to no fine-tuning and demonstrate impressive performance, such a paradigm has yet to succeed in the graph learning domain.
The primary goal of this workshop is to expand the impact of graph learning beyond the current boundaries. Graph learning is poised to become a broadly useful toolbox for learning and understanding any type of (structured) data.
This workshop is presented at the International Conference on Neural Information Processing Systems (NeurIPS).
AI for Science (ICML 2022)
Machine learning is poised to transform scientific discovery. Despite this promise, critical challenges stifle algorithmic and scientific innovation across scientific disciplines.
This workshop brings those challenges to the forefront and discusses which are likely/unlikely to have a broad impact on scientific discovery. With hundreds of AI/ML scientists beginning projects in this area, the workshop brings them together to facilitate community building and consolidate the fast growing area of AI4Science into a vibrant research field.
This workshop is presented at the International Conference on Machine Learning (ICML).
Trustworthy AI for Healthcare (AAAI 2022)
AI for healthcare has emerged into a very active research area in the past few years and has made significant progress. For example, AI methods have achieved human-level performance in skin cancer classification, diabetic eye disease detection, chest radiograph diagnosis, sepsis treatment.
While current results are encouraging, few clinical AI solutions are deployed in hospitals or routinely used in the clinic. The major problem is that existing clinical AI methods are not sufficiently trustworthy. Black-box methods generate decisions that are difficult to understand and interpret. Furthermore, existing solutions are sensitive to small perturbations and adversarial attacks, which raises security and privacy concerns. Further, methods can produce results biased against certain populations. This workshop addresses these challenges and puts forward recommendations on how to make clinical AI solutions more trustworthy.
This workshop was presented at the AAAI Conference on Artificial Intelligence (AAAI 2022).
Workshop on Graph Learning Benchmarks (The Web Conference 2022)
While graph neural networks achieve promising performance on node classification, graph classification, and link prediction, reported performance gains can only be verified and compared within a limited set of publicly available benchmark datasets. The lack of diversity in benchmark datasets may have biased the development of graph representation learning techniques towards narrow directions.
By crowdsourcing tasks and datasets, the workshop has increased the diversity of graph learning benchmarks, identified open questions in graph representation learning, and deepened the synergy between graph ML algorithms and benchmark datasets.
The workshop was organized at the Web Conference (WWW).
AI for Science (NeurIPS 2021)
Machine learning has advanced a wide array of scientific disciplines and addressed many problems that previously could not be tackled computationally. Despite this promise, several key challenges remain open, and this workshop brings those gaps to the foreground of AI research.
-
Gap 1: Unrealistic methodological assumptions. While ML researchers strive for methodology advances, they often make unrealistic assumptions that limit real-world adoption. For example, most state-of-the-art molecule generation ML models generate molecules that have low synthesizability.
-
Gap 2: Overlooked scientific questions. Scientific communities contend with crucial and unsolved problems, but they are not yet formulated as solvable ML tasks and are thus overlooked by the ML community.
-
Gap 3: Limited exploration at the intersection of multiple disciplines. Solutions to grand challenges often stretch across multiple disciplines. For example, protein structure prediction requires collaboration across physics, chemistry and biology.
-
Gap 4: Science of science. Core principles of the scientific method have not changed since the 17th century. Can AI reason about the organizing principles of our world in a way that is complementary to the hypothesis-experiment cycle to understand a phenomenon?
-
Gap 5: Responsible use and development of AI for science. Interest in ML across scientific disciplines has surged, but few ML models have transitioned into practical scientific applications. We plan to present a roadmap and ultimately guidelines for accelerating the translation of ML in science. Translation requires a team of engaged stakeholders and a systematic process from the beginning (problem formulation) to the end (widespread deployment) of ML-based research lifecycle.
This workshop was presented at the International Conference on Neural Information Processing Systems (NeurIPS).
Trustworthy AI for Healthcare (AAAI 2021)
Artificial intelligence for healthcare has emerged as an active research area that has made considerable progress, including achieving human-level performance for skin cancer classification, diabetic eye disease detection, chest radiograph diagnosis, and sepsis treatment. While the trends are encouraging, many open challenges prevent us from directly deploying AI solutions in hospitals and clinical environments. A major open problem is the lack of trust of biomedical practitioners in AI methods. Many AI methods make predictions in a black-box way, making decisions challenging to understand and interpret. Further, today's methods are sensitive to small perturbations and adversarial attacks, raising numerous security and privacy concerns. Finally, AI methods learn to make decisions based on training data, which can include biased human decisions or reflect historical or social inequities. These challenges raise numerous trustworthy issues that we need to address to realize the potential of AI in healthcare.
This workshop was presented at the AAAI Conference on Artificial Intelligence (AAAI 2021).
AI in Health: Transferring and Integrating Knowledge for Better Health (The Web Conference 2021)
Rich healthcare data connected by semantic relationships and integrated into knowledge graphs can drive biomedical discovery. Biomedical knowledge graphs can support better cohort identification for clinical trials, risk prediction, precision diagnosis, and can inform new and better decision support workflows. Dramatic increase of healthcare data offers unprecedented opportunities for evidence-based care, yet challenges related to interoperability, learning, and reasoning over healthcare data remain open.
This workshop was presented at the Web Conference (WWW).
National Symposium on Drug Repurposing for Future Pandemics (2020)
Pandemics demand safe and effective therapies developed and deployed at an unprecedented speed. This symposium, organized on behalf of the National Science Foundation (NSF), provides a forum for scientists and researchers from a variety of fields relevant to therapeutics. Participants discuss ways to expedite the development of therapies by compressing years of work into months or even weeks through automation, artificial intelligence and machine learning, novel data sources, and most recent biotechnology advancements.

The symposium brings together leading experts in computer science, biology, statistics, medicine, automation, and regulation. While these areas of expertise are necessary for rapid therapeutic innovation, there is seldom an opportunity for these experts to interact with each other.
Bearing in mind new opportunities and pressing challenges, the symposium provides a roadmap and put forward recommendations on transforming today’s tools into ready-to-use solutions to fight future pathogens.
We announce a new initiative, Therapeutics Data Commons (TDC), at the symposium [Slides].

Graph Representation Learning and Beyond (ICML 2020)
Recent years have seen a surge in research on graph representation learning, including techniques for deep graph embeddings, generalizations of CNNs to graph-structured data, and neural message-passing approaches. These advances in graph neural networks and related techniques have led to new state-of-the-art results in numerous domains: chemical synthesis, 3D-vision, recommender systems, question answering, continuous control, self-driving, and social network analysis.
This workshop was presented at the International Conference on Machine Learning (ICML).
Representation Learning on Graphs and Manifolds (ICLR 2019)
Many scientific fields study data with an underlying graph or manifold structure—such as social networks, sensor networks, biomedical knowledge graphs, and meshed surfaces in computer graphics. Recent years have seen a surge in research on these problems—often under the umbrella terms of graph representation learning and geometric deep learning.
The need for new optimization methods and neural network architectures that can accommodate these relational and non-Euclidean structures is becoming increasingly clear. In parallel, there is a growing interest in how we can leverage insights from these domains to incorporate new kinds of relational and non-Euclidean inductive biases into deep learning.
This workshop was presented at the International Conference on Learning Representations (ICLR).
Research and Scholarship Meetings
Therapeutics Data Commons User Group Meeting (2022)
Therapeutics Data Commons is an open-science initiative started at Harvard with AI/ML-ready datasets and ML tasks for therapeutics. TDC provides an ecosystem of tools, leaderboards, and community resources, including data functions, strategies for model benchmarking and comparison, meaningful data splits, data processors, public leaderboards, and molecule generation oracles. All resources are integrated via an [open Python library.](https://github.com/mims-harvard/TDC)
The lack of high-quality benchmarks impedes the advancement of ML tools for drug discovery. To this end, TDC supports the development of novel ML theory and methods, with a strong bent towards developing the mathematical foundations of which ML algorithms are most suitable for drug discovery applications and why. TDC contains benchmarks for therapeutics ML tasks, including molecular property prediction, molecular interaction prediction, and molecular optimization, all accompanied by extensive programmatic support and leaderboards.
The first live user meeting outlined opportunities for how to engage with the TDC community, provided technical background and demos on how TDC supports ML for therapeutics and molecules.
PhD Forum (ECML/PKDD 2020)
PhD Forum provides an environment for junior PhD students to exchange ideas and experiences with peers in an interactive atmosphere and to get constructive feedback from senior researchers in data science, machine learning, and related areas.
This meeting took place at the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML-PKDD).
Seminar Series
Harvard University Efficient ML Seminar Series
With the advent of foundation models and the emergence of capabilities at scale, algorithms designed to improve the training and inference efficiency of machine learning systems have become critical research topics. Despite their importance, expertise in training the most efficient systems is often in short supply, limiting the ability of teams to scale experiments, work at maximum productivity, develop foundation models for underexplored domains, and make the best use of our computing resources.
This seminar series seeks to educate, provide a platform for, and facilitate collaborative research opportunities for all algorithmic and technological questions arising in the quest for more efficient machine learning at scale:
-
A proliferation of knowledge on state-of-the-art approaches directly employable by diverse research teams hoping to scale training or make use of large-scale models.
-
The provision of a research discussion forum on emerging topics to help catalyze further research and form connections among researchers invested in researching these questions.
Research Tutorials
Towards Precision Medicine with Graph Representation Learning (ISMB 2022)
Graph representation learning has matured immensely as a field within the last few years. Graph machine learning approaches, also known as geometric deep learning, or graph neural networks has become widely used in biomedical applications. This tutorial surveys impact areas in precision medicine (e.g., modeling disease progression, candidate biomarker discovery for targeted therapies, rapid disease diagnostics, treatment regimen recommendations) and highlights new opportunities enabled by these approaches.
This tutorial was presented at the International Conference on Intelligent Systems for Molecular Biology (ISMB).
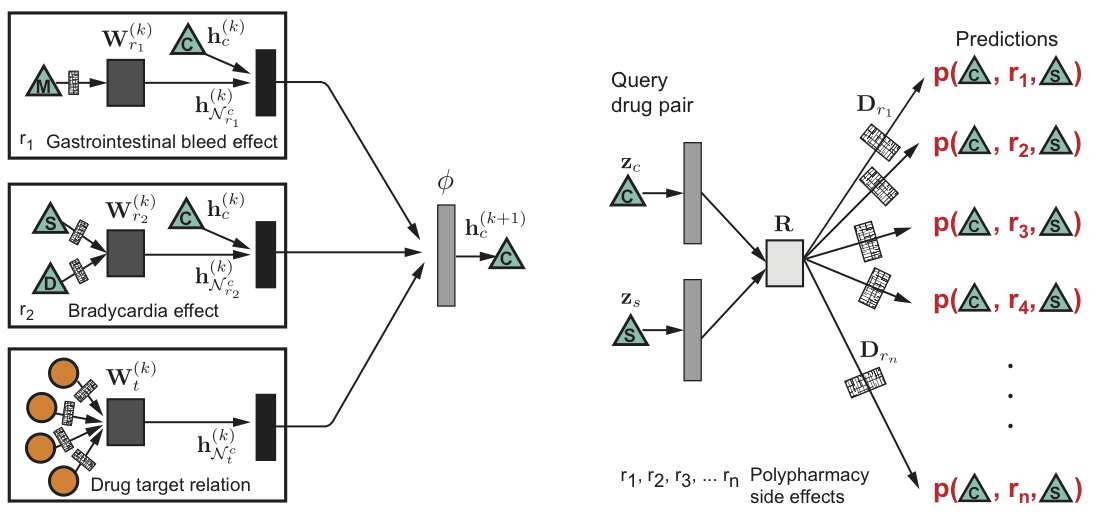
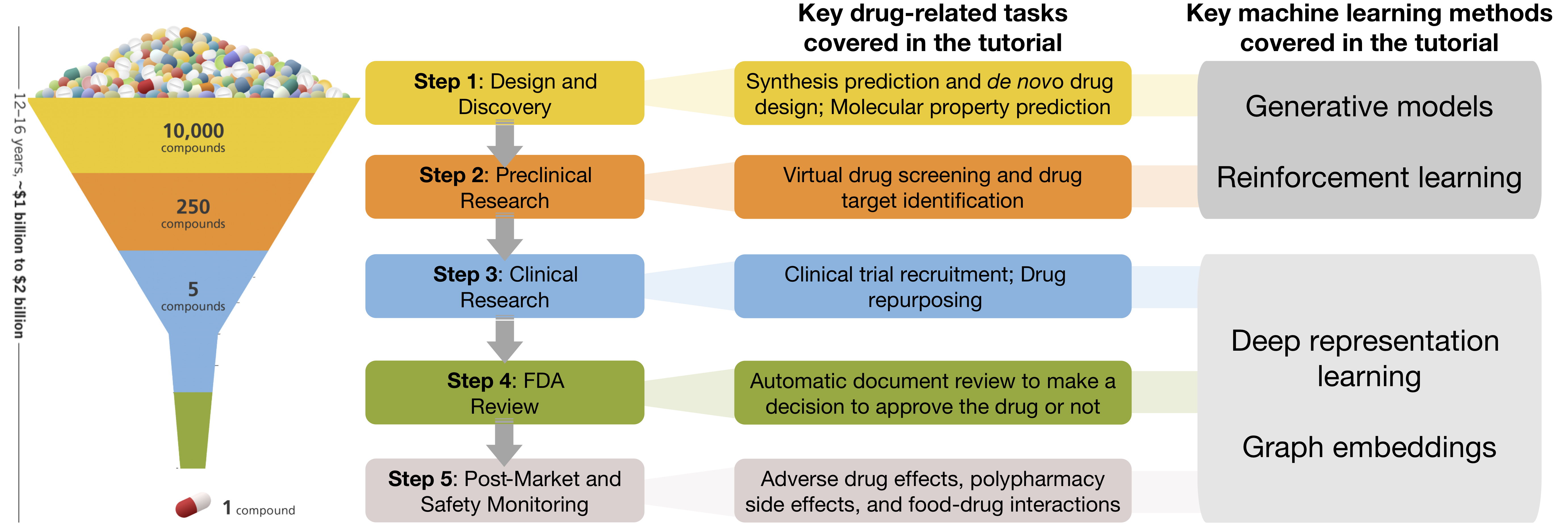
Machine Learning for Drug Development (IJCAI 2021)
Machine learning methods leverage big datasets to support decision-making in all stages of drug development, predict how drugs affect the human body and how they interact with each other, and seek ways to boost clinical trials and detect unwanted side effects. This tutorial covers generative modeling, reinforcement learning, and representation learning with a focus on theoretical foundations of methods and their use for key drug-related problems.
A variety of machine learning methods are demonstrating their utility at all stages of drug development. These methods use big datasets created from high-throughput screening data and allow prediction of bioactivities for targets and molecular properties, identification of new molecules and repurposing of old drugs with increased levels of accuracy.
We have only just begun to realize the potential of these techniques. If methods were available for all aspects of drug development, they could be used seamlessly to predict whether a chemical compound is likely to ultimately become a drug used in patients. Much research needs to be done before this vision can be realized, modern machine learning may have a fundamental impact on the way drug development is done.
The general process of drug development involves five steps. In short, molecular compounds are filtered through a progressive series of tests, which determine their properties, toxicity, and effectiveness for later stages. Machine learning is increasingly being used to accelerate each of the steps, creating opportunities for reducing resources and time needed to develop new drugs. In this tutorial, we cover key problems in drug development that are amenable to machine learning. In doing so, we present a toolbox of AI algorithms for end-to-end drug development.
This tutorial was presented at the International Joint Conference on Artificial Intelligence (IJCAI).
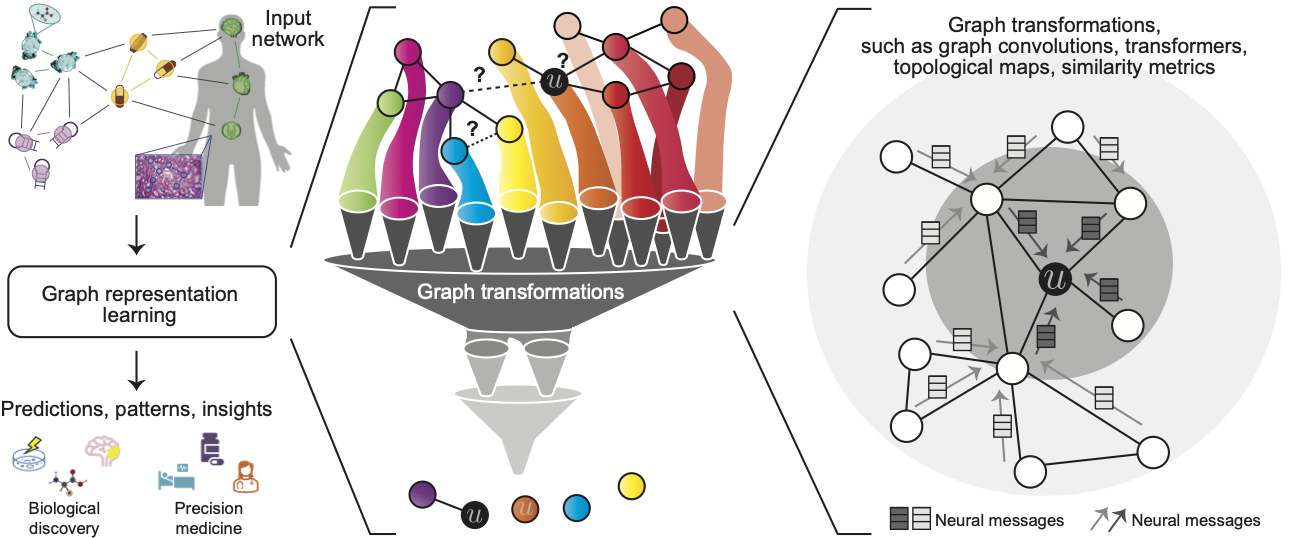
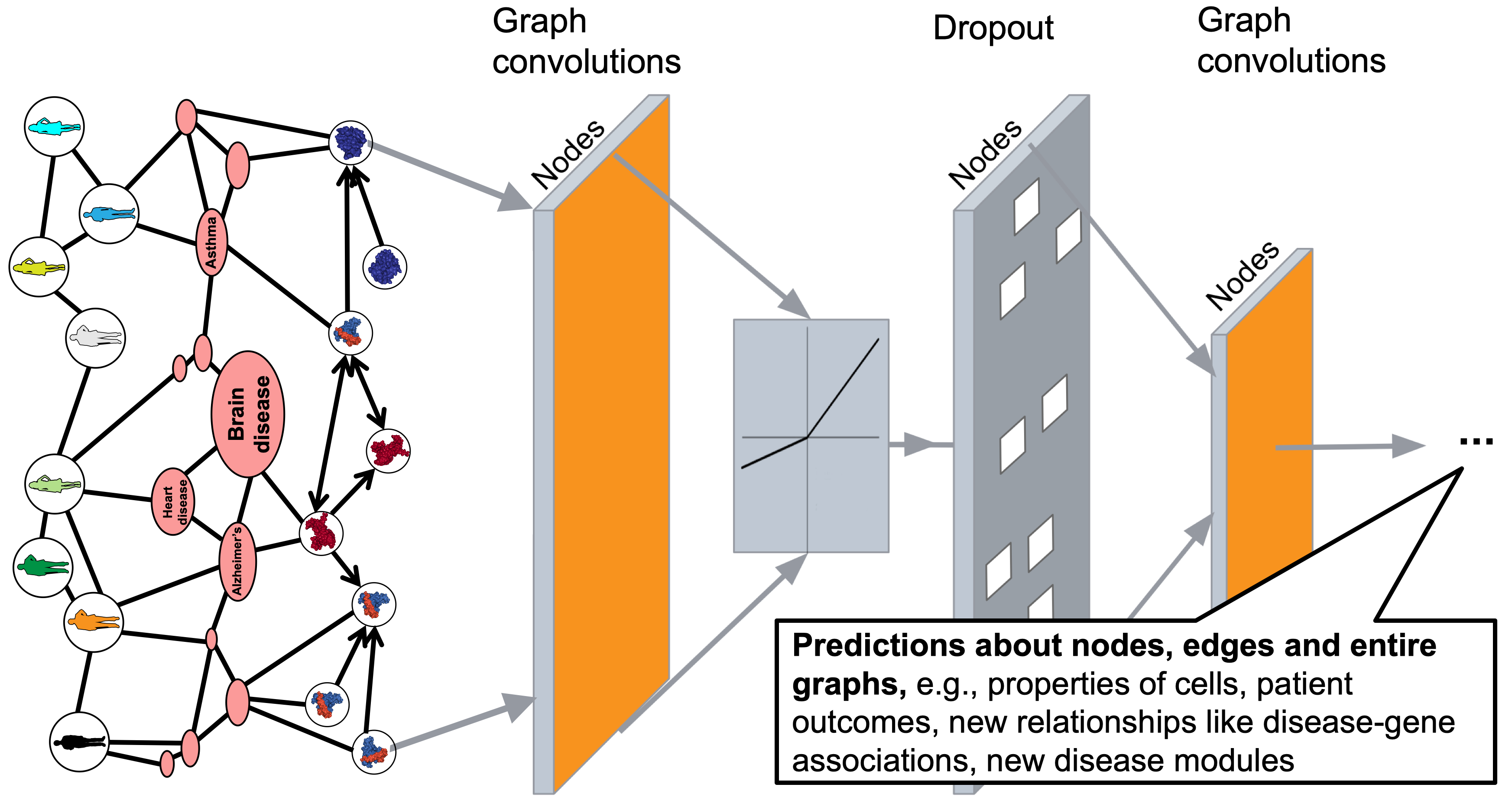
Deep Learning for Network Biology (ISMB 2018)
Networks are ubiquitous in biology where they encode connectivity patterns at all scales of organization, from molecular to the biome. This tutorial investigates key advancements in representation learning for networks over the last few years, with an emphasis on fundamentally new opportunities in network biology enabled by these advancements.
Biological networks are powerful resources for the discovery of interactions and emergent properties in biological systems, ranging from single-cell to population level. Network approaches have been used many times to combine and amplify signals from individual genes, and have led to remarkable discoveries in biology, including drug discovery, protein function prediction, disease diagnosis, and precision medicine. Furthermore, these approaches have shown broad utility in uncovering new biology and have contributed to new discoveries in wet laboratory experiments.
Mathematical machinery that is central to these approaches is machine learning on networks. The main challenge in machine learning on networks is to find a way to extract information about interactions between nodes and to incorporate that information into a machine learning model. To extract this information from networks, classic machine learning approaches often rely on summary statistics (e.g., degrees or clustering coefficients) or carefully engineered features to measure local neighborhood structures (e.g., network motifs). These classic approaches can be limited because these hand-engineered features are inflexible, they often do not generalize to networks derived from other organisms, tissues and experimental technologies, and can fail on datasets with low experimental coverage.
Recent years have seen a surge in graph neural network (GNN) approaches that automatically learn to encode network structure into low-dimensional representations, using transformation techniques based on deep learning and nonlinear dimensionality reduction. The idea behind these representation learning approaches is to learn a data transformation function that maps nodes to points in a low-dimensional vector space, also termed embeddings. Representation learning methods have revolutionized the state-of-the-art in network science and the goal of this tutorial is to open the door for these methods to computational biology and bioinformatics.
This tutorial was presented at the International Conference on Intelligent Systems for Molecular Biology (ISMB).
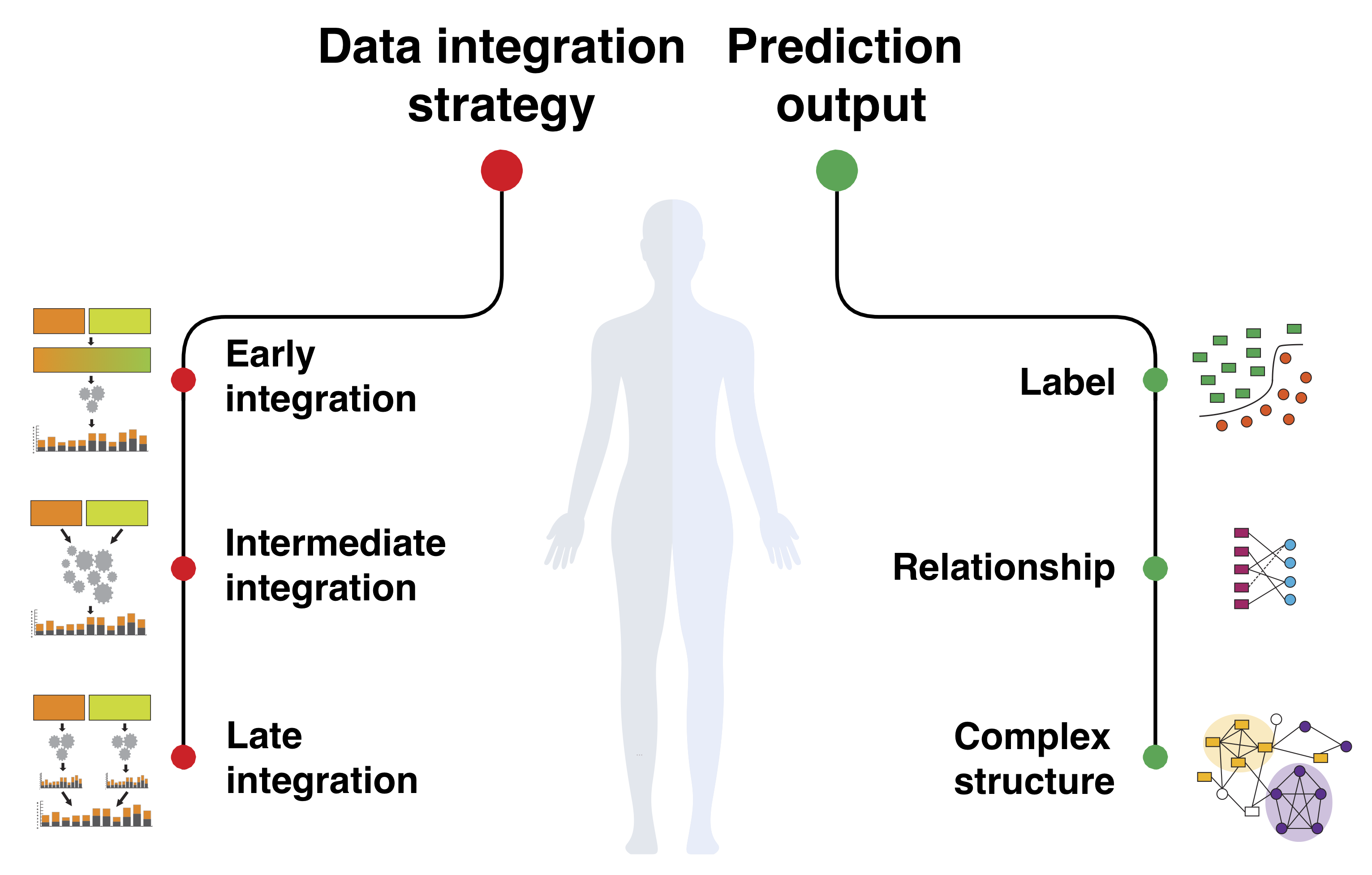
Biomedical Data Fusion (EMBC and [BC]^2 2015)
Because of the complex and interconnected nature of biomedical systems, any single model trained on any single dataset can touch only a small part of the entire biomedical knowledge. It is thus critical to integrate diverse sources of information to gain a comprehensive understanding of the system.
New technologies have enabled the investigation of biology and human health at an unprecedented scale and in multiple dimensions. These dimensions include a myriad of properties describing genome, epigenome, transcriptome, microbiome, phenotype, and lifestyle. No single data type, however, can capture the complexity of all the factors relevant to understanding a phenomenon such as a disease. Integrative methods that combine data from multiple technologies have thus emerged as critical statistical and computational approaches.
The key challenge in developing such approaches is the identification of effective models to provide a comprehensive and relevant systems view. An ideal method can answer a biological or medical question, identifying important features and predicting outcomes, by harnessing heterogeneous data across several dimensions of biological variation.
This tutorial was presented at the International Engineering in Medicine and Biology Conference (EMBC) and at the Basel Compuational Biology Conference ([BC]^2).