Foundation models show promise in medicine, but clinical use requires outputs that clinicians can act on under pre-specified error budgets, such as a cap on false-positive clinical calls. Without error control, strong average accuracy can still lead to harmful errors among the very cases labeled confident and to inefficient use of follow-up testing.
Here we introduce StratCP, a stratified conformal framework that turns foundation-model predictions into decision-ready outputs by combining selective action with calibrated deferral. StratCP first selects a subset of patients for immediate clinical calls while controlling the false discovery rate among those calls at a user-chosen level. It then returns calibrated prediction sets for deferred patients that meet the target error rate and guide confirmatory evaluation. The procedure is model agnostic and can be applied to pretrained foundation models without retraining.
We validate StratCP in ophthalmology and neuro-oncology across diagnostic classification and time-to-event prognosis. Across tasks, StratCP maintains false discovery rate control on selected patients and produces coherent prediction sets for deferred patients. In neuro-oncology, it supports diagnosis from H&E whole-slide images under a fixed error budget, reducing the need for reflex molecular assays and lowering laboratory cost and turnaround time. StratCP lays the groundwork for safe use of medical foundation models by converting predictions into error-controlled actions when evidence is strong and calibrated uncertainty otherwise.
Overview of StratCP
Foundation models (FMs) perform well across clinical tasks, including retinal imaging, whole-slide histopathology, and clinical question answering. Some FMs have also been tested in prospective or health system settings, including language models evaluated on diagnosis and risk prediction tasks and a pathology FM assessed in silent use for EGFR screening. Routine clinical use, however, shifts the requirement from performing well on benchmarks to producing outputs that support safe action under pre-specified error budgets. The key challenge is not only how accurate a model is on average, but when it is appropriate to act on its output. Clinicians need guidance on when to make a call, when to defer, and what follow up to order. This requires decision policies with pre-specified error budgets. Without them, model driven decisions can trigger unnecessary or harmful interventions, delay appropriate care, and waste limited diagnostic resources.
For safe clinical use of foundation models, uncertainty quantification should provide two guarantees. First, it should identify cases where a prediction is reliable enough to act on. Second, it should return calibrated candidate diagnoses for the remaining cases to guide confirmatory testing or expert review. The first requires error control within the acted upon selected subset. The second requires coverage within the deferred group. With guarantees aligned to clinical decisions, a clinician can make a call under a specified error budget or defer with calibrated differential diagnosis sets, enabling safe action when evidence is sufficient and follow up when it is not.
To meet these guarantees, we introduce StratCP, a stratified conformal framework that combines selective action with calibrated deferral and can wrap any trained FM without retraining.
-
In the action arm, StratCP selects a subset of cases for immediate decisions while controlling the false-discovery rate (FDR), the expected fraction of incorrect acted-upon predictions, at a pre-specified level for task-specific outputs, such as tumor subtype or long-survivor status.
-
In the deferral arm, StratCP returns conformal prediction sets with selection-conditional coverage. This ensures that, among deferred patients, the true diagnosis falls within the set at the target frequency.
When available, StratCP can use clinical diagnostic guidelines to produce clinically coherent prediction sets without sacrificing coverage.
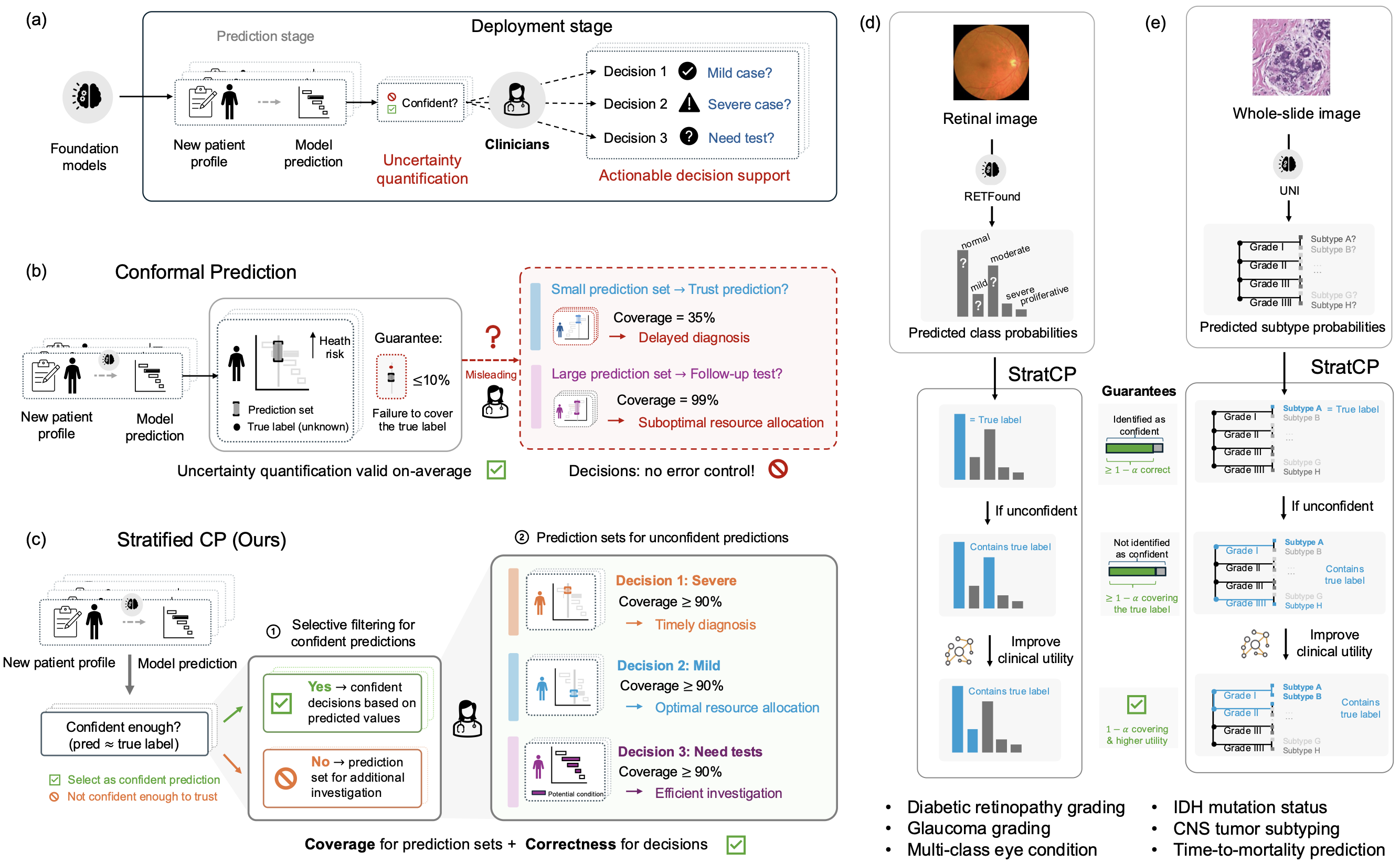
StratCP Provides Guarantees for Safe Medical Decisions
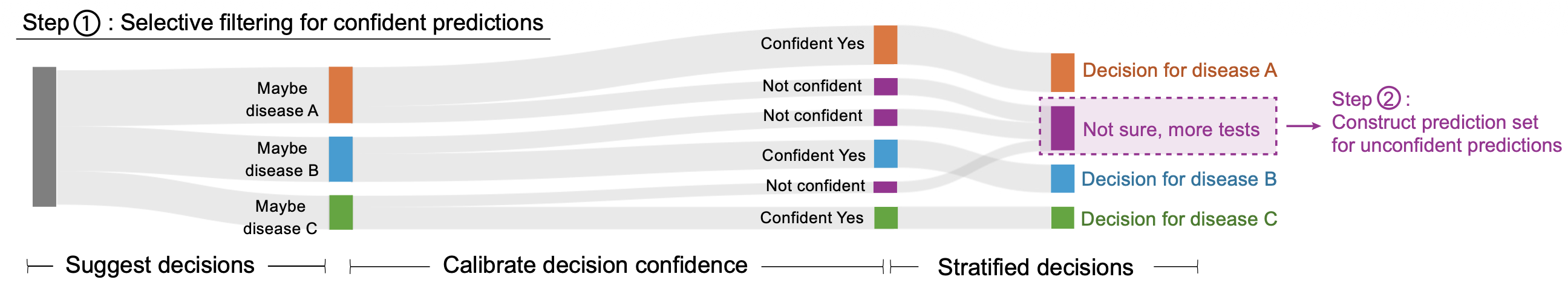
StratCP builds on conformal prediction, a widely used approach for uncertainty quantification. Given a specified error tolerance, such as 5%, and a set of new patients, StratCP proceeds in two steps. First, it selects cases where the model prediction can be used for an immediate call, while keeping the error rate among these acted-upon cases below the pre-specified budget (for example, fewer than 5% of selected predictions are incorrect). Second, for cases not selected for an immediate call, StratCP returns a prediction set of plausible labels, such as normal, mild, with a guarantee that the true disease status lies in the set for 95% of deferred patients (see Methods for details and theoretical guarantees).
Step 1: Select confident predictions under an error budget: StratCP turns the model output into candidate decisions, such as assigning a specific disease status. For each candidate diagnosis, StratCP uses expert-labeled reference data to compute a confidence measure for each new patient and sets a decision threshold by testing against the reference data. Cases whose confidence exceeds this threshold are marked as confident. These confident calls can then be used directly for decision making under the chosen error budget.
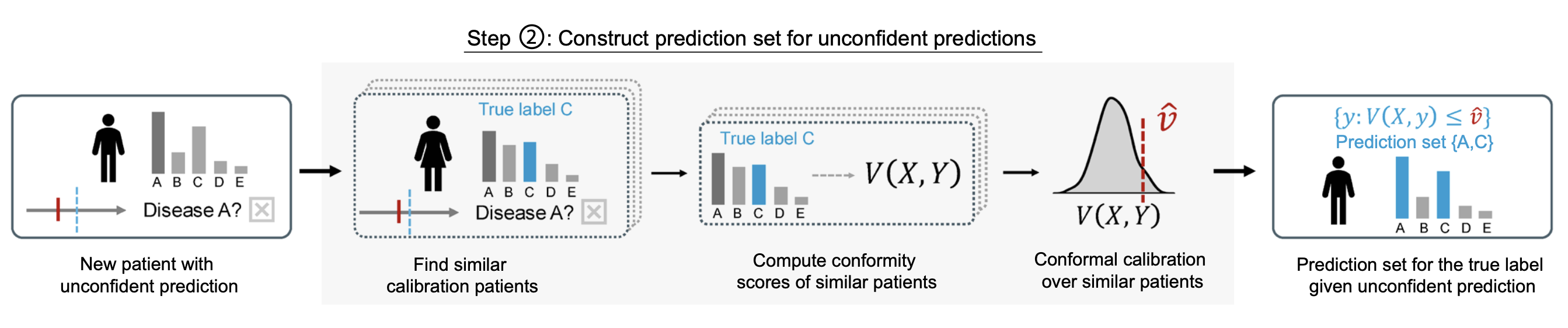
Step 2: Calibrated prediction sets for deferred cases: The second step of StratCP returns prediction sets for patients whose FM-predicted diagnoses are not confident enough for an immediate call. After Step 1, deferred cases differ from the full reference set because they are enriched for ambiguity. StratCP accounts for this shift by calibrating uncertainty using only expert-labeled cases with similar ambiguity, following recent work on post-selection conformal inference. Given an unconfident case, StratCP finds a subset of expert-diagnosed patients whose FM outputs are similarly ambiguous, using the same confidence criterion as in Step 1. It then forms a prediction set by including each candidate diagnosis for which the corresponding test statistic falls within the expected range when compared to these similarly ambiguous expert-labeled cases.
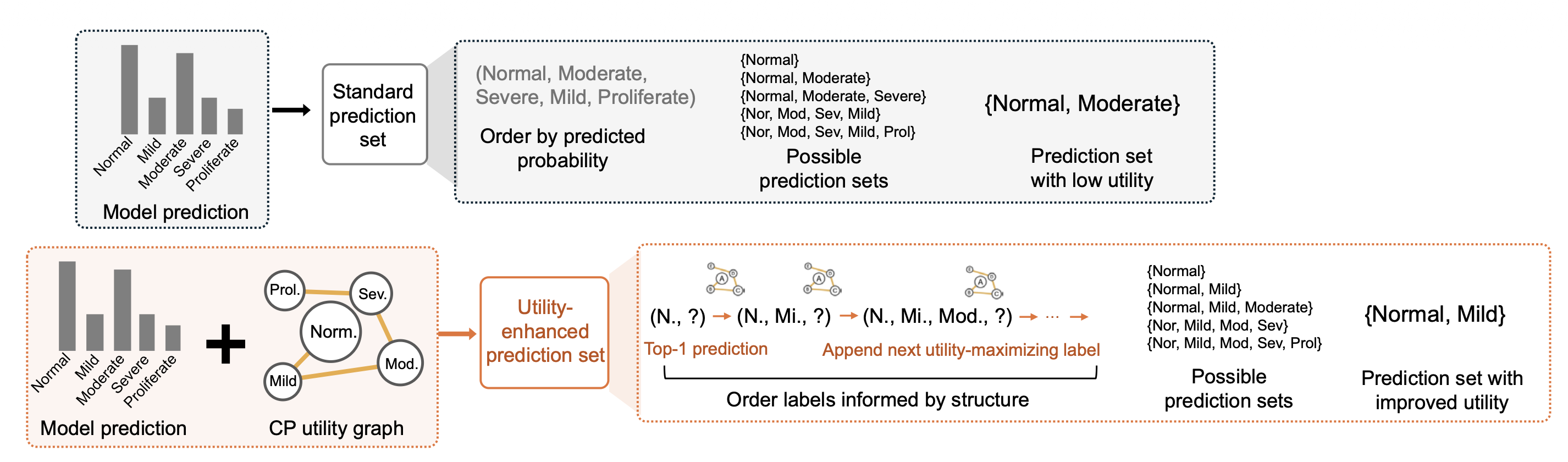
Optional utility enhancement using diagnostic-guideline knowledge: When diagnostic-guideline knowledge about relationships among labels is available, StratCP includes an optional module that shapes prediction sets to better match clinical reasoning for deferred cases. StratCP changes the order in which candidate labels enter the prediction set by combining model scores with a CP utility graph derived from diagnostic guidelines that encodes which label combinations are preferred. Starting from the most probable model label, it iteratively adds the label that increases set utility the most, until the set reaches the size required for valid coverage.
Safe Use of Medical Foundation Models With StratCP
We evaluate StratCP in ophthalmology and oncology, pairing it with a vision foundation model on retinal images and a pathology foundation model on H&E whole-slide images for diagnosis, biomarker prediction, and prognosis. StratCP controls error among acted-upon (selected) cases and returns valid coverage for deferred cases, so the outputs map to decision points.
In eye-condition diagnosis, both StratCP and CP meet 95% coverage, but StratCP yields more actionable calls on average while keeping the selected-set error rate near the target.
In IDH mutation status prediction, StratCP keeps the acted-upon error rate within the 5% budget for IDH-mutant calls (FDR 0.046), whereas a standard conformal prediction approach overspends the budget on its acted-upon subset (FDR 0.110).
In time-to-event prognosis, StratCP reaches the nominal 95% coverage among selected long-survivors (0.952), while a threshold baseline falls short (0.797).
We also show that utility graphs derived from expert diagnostic guidelines can shape prediction sets without sacrificing conformal coverage. This produces differential diagnosis sets that respect clinical adjacency, such as neighboring diabetic retinopathy stages or related CNS tumor categories, and better match the follow-up actions a clinician would take.
Finally, in a neuro-oncology application, StratCP provides error-controlled diagnoses for adult-type diffuse glioma. By finalizing a subset of cases without reflex molecular testing within the error budget, it can reduce confirmatory assays, lowering laboratory cost and shortening time to diagnosis.
Publication
Error Controlled Decisions for Safe Use of Medical Foundation Models
Ying Jin*, Intae Moon*, Marinka Zitnik
In Review 2026 [arXiv]
@article{jin26error,
title={Error Controlled Decisions for Safe Use of Medical Foundation Models},
author={Jin, Ying and Moon, Intae and Zitnik, Marinka},
journal={In Review},
url={},
year={2026}
}
Code and Data Availability
Pytorch implementation of StratCP is available in the GitHub repository.