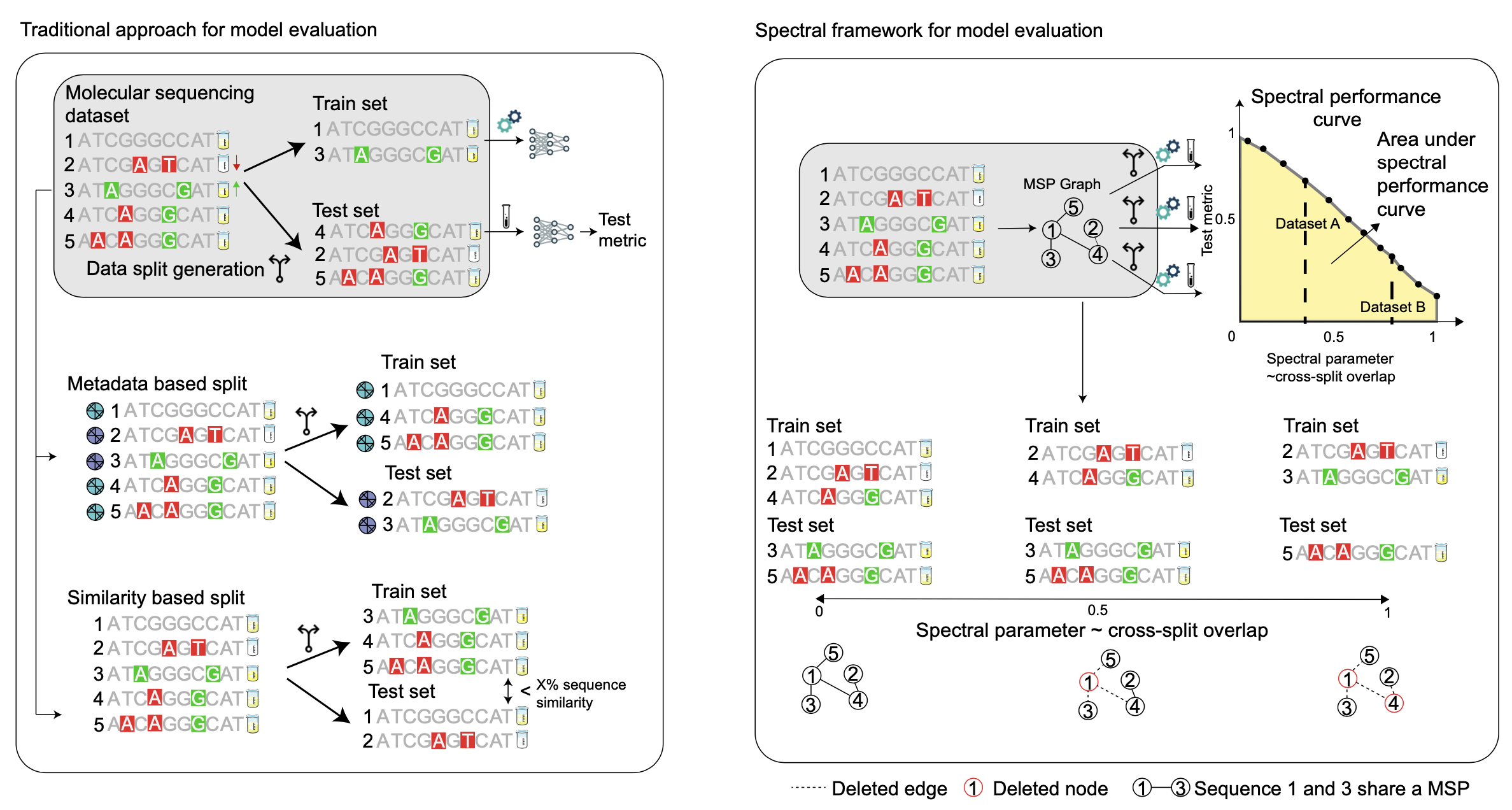
Deep learning has made rapid advances in modelling molecular sequencing data. Despite achieving high performance on benchmarks, it remains unclear to what extent deep learning models learn general principles and generalize to previously unseen sequences. Benchmarks traditionally interrogate model generalizability by generating metadata- or sequence similarity-based train and test splits of input data before assessing model performance.
Here we show that this approach mischaracterizes model generalizability by failing to consider the full spectrum of cross-split overlap, that is, similarity between train and test splits. We introduce SPECTRA, the spectral framework for model evaluation. Given a model and a dataset, SPECTRA plots model performance as a function of decreasing cross-split overlap and reports the area under this curve as a measure of generalizability.
We use SPECTRA with 18 sequencing datasets and phenotypes ranging from antibiotic resistance in tuberculosis to protein–ligand binding and evaluate the generalizability of 19 state-of-the-art deep learning models, including large language models, graph neural networks, diffusion models and convolutional neural networks. We show that sequence similarity- and metadata-based splits provide an incomplete assessment of model generalizability.
Using SPECTRA, we find that as cross-split overlap decreases, deep learning models consistently show reduced performance, varying by task and model. Although no model consistently achieved the highest performance across all tasks, deep learning models can, in some cases, generalize to previously unseen sequences on specific tasks. SPECTRA advances our understanding of how foundation models generalize in biological applications.
Publication
Evaluating Generalizability of Artificial Intelligence Models for Molecular Datasets
Yasha Ektefaie, Andrew Shen, Daria Bykova, Maximillian Marin, Marinka Zitnik* and Maha Farhat*
Nature Machine Intelligence 2024 [bioRxiv]
@article{ektefaie2024evaluating,
title={Evaluating Generalizability of Artificial Intelligence Models for Molecular Datasets},
author={Ektefaie, Yasha and Shen, Andrew and Bykova, Daria and Maximillian, Marin and Zitnik, Marinka* and Farhat, Maha*},
journal={Nature Machine Intelligence},
url={https://rdcu.be/d2D0z},
year={2024}
}
Code Availability
Pytorch implementation of SPECTRA is available in the GitHub repository.