When we train a model to recognize patterns in one type of data, it might not work well when we apply it to a different type of data. This is where unsupervised domain adaptation (UDA) comes in, which helps us transfer the trained model to a different type of data without needing labeled examples.
However, when dealing with complex time series data (like sensor readings over time), it becomes harder to transfer the model because the patterns in the data change over time and can be different between different domains (like different sensors or environments). Additionally, the labels that we use to train the model might be different between domains. This means that the model might not work well when we apply it to the new domain.
To address this problem, a new model Raincoat can handle complex time series data by aligning the different patterns in the data across domains and correcting for any differences. It can also detect when the labels are different between domains and adjust the model accordingly.
Motivation
Training models that can adapt to domain shifts is crucial for robust, real-world implementation. We often encounter data from different sources or domains, which can lead to issues with model performance. That is where domain adaptation comes in – it is all about creating models that can adapt to different domains and perform well in any setting. For example, in healthcare, time series data collection methods vary widely across clinical sites, leading to shifts in underlying features and labels. Endowing learning systems with domain adaptation capabilities increases their reliability and expands applicability across downstream tasks.
However, building models that can adapt to different domains is no easy feat. There are a variety of challenges, including feature and label shifts, private labels, and more. Domain adaptation is a highly complex problem due to several factors, including private labels that do not exist in the source domain. Models trained for robustness to domain shifts must learn highly generalizable features to avoid relying on spurious correlations created by non-causal data artifacts. Shifts in label distributions across domains may result in private labels, making domain adaptation challenging. Traditional techniques often fall short when it comes to these challenges, and that is where new, innovative approaches are needed.
One approach to domain adaptation is adversarial learning, where a model is trained to be invariant to differences between the source and target domains. Another is transfer learning, where a model is pre-trained on a large dataset and then fine-tuned on a smaller, domain-specific dataset. But perhaps the most exciting approach to domain adaptation is unsupervised learning, where a model is trained on data from the source domain and then applied to the target domain without any labeled data. This approach has enormous potential for real-world applications, as it does not require labeled data from every domain.
Raincoat
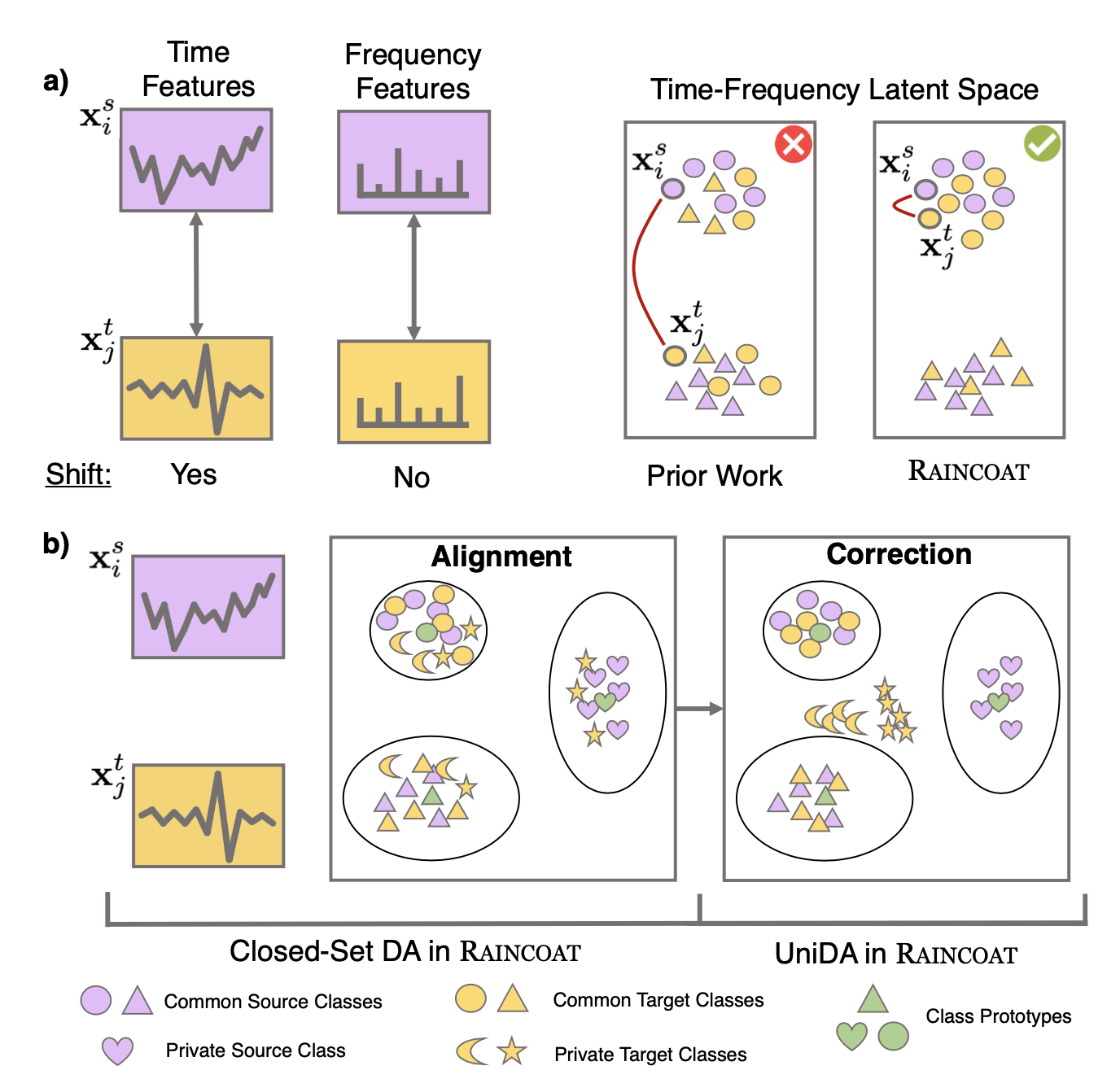
We introduce Raincoat, a domain adaptation method for time series that can handle both feature and label shifts. Raincoat is the first model for both closed-set and universal domain adaptation on complex time series. It addresses feature and label shifts by considering both temporal and frequency features, aligning them across domains, and correcting for misalignments to detect private labels.
Raincoat improves transferability by identifying label shifts in target domains. It produces generalizable representations robust to feature and label shifts, expanding the scope of existing domain adaptation methods.
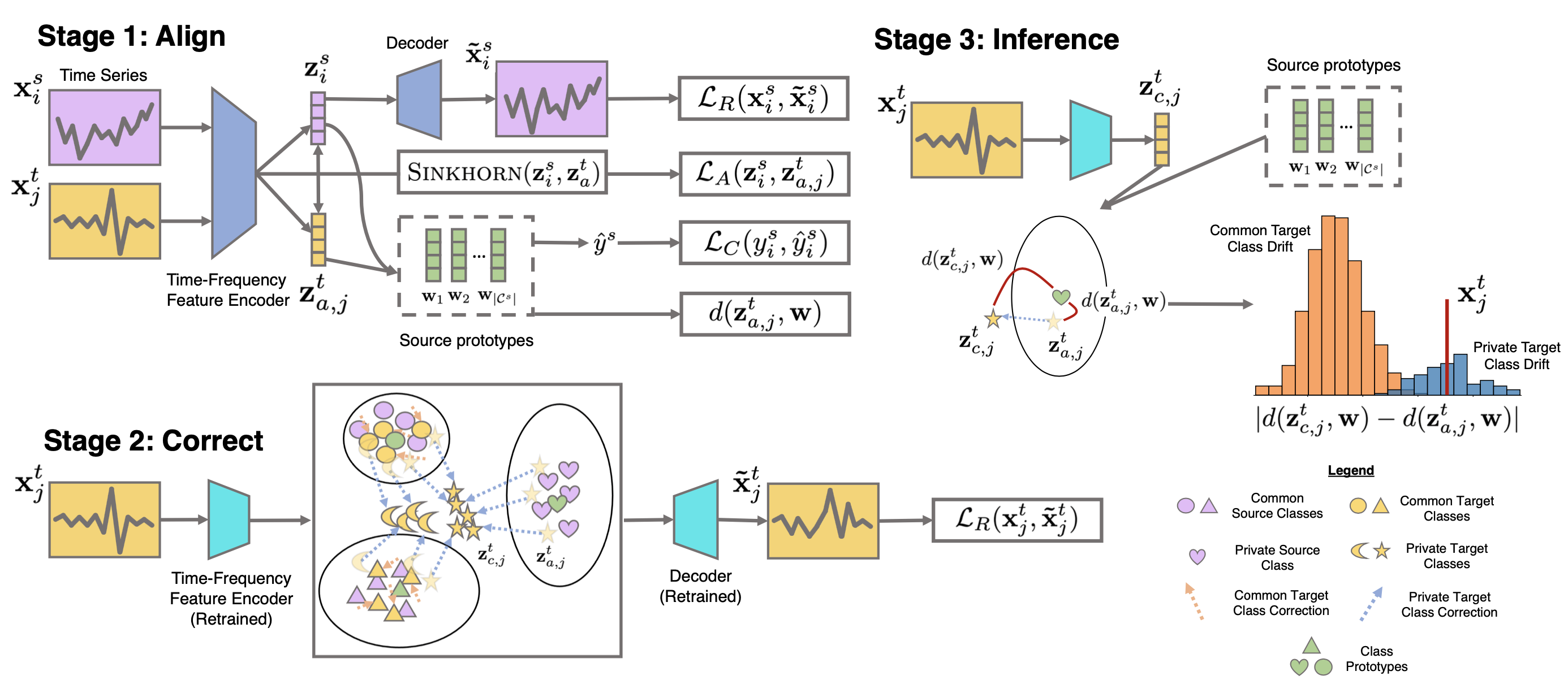
Raincoat Model
Raincoat has three steps, as illustrated in the following figure:
-
Step 1: Align - It employs time and frequency-based encoders to learn time series representations, using Sinkhorn divergence for source-target feature alignment as frequency features may not have the same support.
-
Step 2: Correct - It retrains an encoder on the target domain to correct any potential misalignments.
-
Step 3: Inference - It calculates the difference between the aligned and corrected representations of target samples to identify unknown target samples through a bi-modality test and binary classification task.
Properties of Raincoat
To address feature shift, Raincoat takes into account implicit frequency feature shift and incorporates additional frequency feature inductive bias in the encoder, to uncover potential invariant features across domains and enhance transferability.
To address label shifts, it employs target-specific feature encoders that retain the semantic meaning of the target domain, enabling inference without relying on user-specified input.
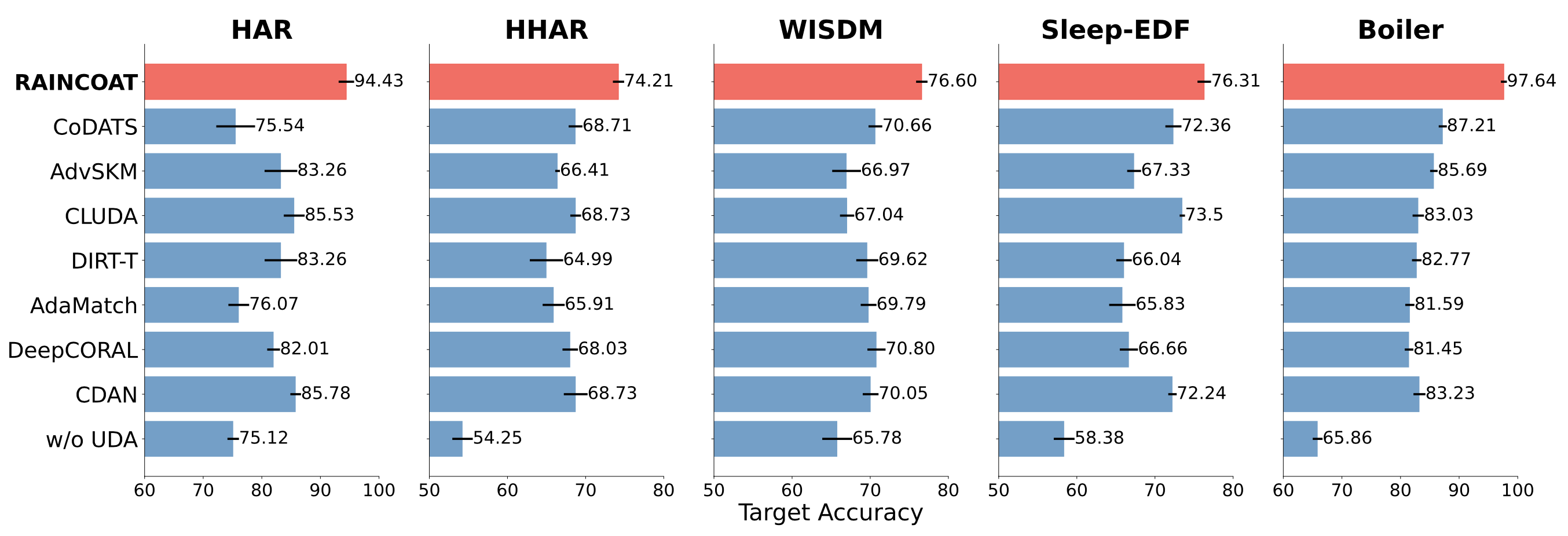
Our experiments using 5 datasets and comparing with 13 state-of-the-art domain adaptation methods show that Raincoat outperforms these methods in the presence of both feature and label shifts. The following figure illustrates the superiority of Raincoat for closed-set domain adaptation.
The results from testing Raincoat on different datasets show that it can improve the performance of the model by up to 16.33%. This means that Raincoat has the potential to be very useful for transferring models between different domains and making them more accurate.
Publication
Domain Adaptation for Time Series Under Feature and Label Shifts
Huan He, Owen Queen, Teddy Koker, Consuelo Cuevas, Theodoros Tsiligkaridis, Marinka Zitnik
International Conference on Machine Learning, ICML 2023 [arXiv] [ICML 2023]
@inproceedings{he2023domain,
title = {Domain Adaptation for Time Series Under Feature and Label Shifts},
author = {He, Huan and Queen, Owen and Koker, Teddy and Cuevas, Consuelo and Tsiligkaridis, Theodoros and Zitnik, Marinka},
booktitle = {International Conference on Machine Learning, ICML},
year = {2023}
}
Datasets
- Human activity recognition datasets:
- Polysomnograms of healthy subjects and of subjects with mild difficulty falling asleep and expert annotations of sleep stages
Code
PyTorch implementation together with documentation and examples of usage is available in the GitHub repository.