Protein representation learning has advanced rapidly with the scale-up of sequence and structure supervision, but most models still encode proteins either as per-residue token sequences or as single global embeddings. This overlooks a defining property of protein organization: proteins are built from recurrent, evolutionarily conserved substructures that concentrate biochemical activity and mediate core molecular functions. Although substructures such as domains and functional sites are systematically cataloged, they are rarely used as training signals or representation units in protein models.
We introduce Magneton, an environment for developing substructure-aware protein models. Magneton provides (1) a dataset of 530,601 proteins annotated with over 1.7 million substructures spanning 13,075 types, (2) a training framework for incorporating substructures into existing protein models, and (3) a benchmark suite of 13 tasks probing representations at the residue, substructural, and protein levels. Using Magneton, we develop substructure-tuning, a supervised fine-tuning method that distills substructural knowledge into pretrained protein models.
Across state-of-the-art sequence- and structure-based models, substructure-tuning improves function prediction, yields more consistent representations of substructure types never observed during tuning, and shows that substructural supervision provides information that is complementary to global structure inputs. The Magneton environment, datasets, and substructure-tuned models are all openly available
Building Substructure into Protein Encoding Models
Proteins are organized into recurrent, modular substructures that provide a natural multiscale vocabulary for their representation. At the finest level are amino acids, which assemble into secondary structure elements such as alpha helices and beta sheets; these in turn combine into higher-order motifs and domains such as beta barrels and zinc fingers. These substructures are responsible for core molecular functions of proteins, such as coordinating metal ions for reaction catalysis or binding to other proteins as parts of cellular signaling networks, and their importance is underscored by their occurrence in proteins across the tree of life.
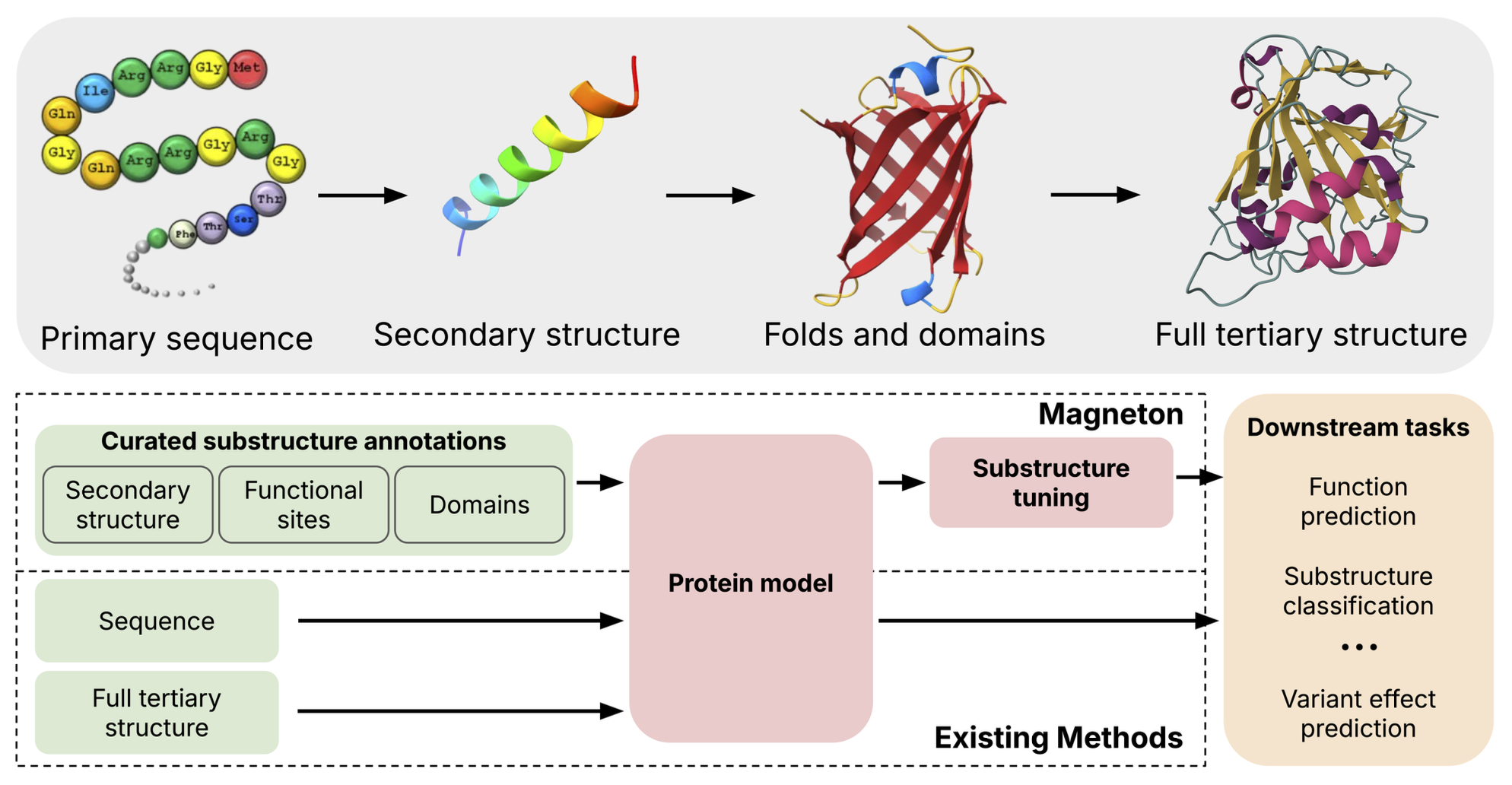
Decades of biological research has led to the categorization of these recurrent substructures, resulting in large databases that exhaustively annotate these elements across proteins. However, prevailing protein representation learning methods still rely on self-supervised objectives that operate at the scale of single amino acids, such as masked language modeling or structural denoising, or occasionally operate on full proteins. This is despite abundant evidence that evolutionarily conserved substructures are key components of protein function. In this work, we ask, how should we systematically incorporate decades of biological knowledge about protein substructures into protein encoding models?
Magneton: Substructure Tuning
Using Magneton, we explore substructure-tuning, a supervised fine-tuning strategy that distills substructural information into protein encoders. Concretely, we formulate substructure-tuning as classification of evolutionarily conserved substructures, where residue-level embeddings produced by a base encoder are pooled to construct substructure representations and optimized with a cross-entropy loss. This objective is model-agnostic, requiring only residue-level embeddings, and naturally extends to multiple structural scales through a multi-task formulation in which each substructure class is assigned its own prediction head and the total loss is the sum across scales.
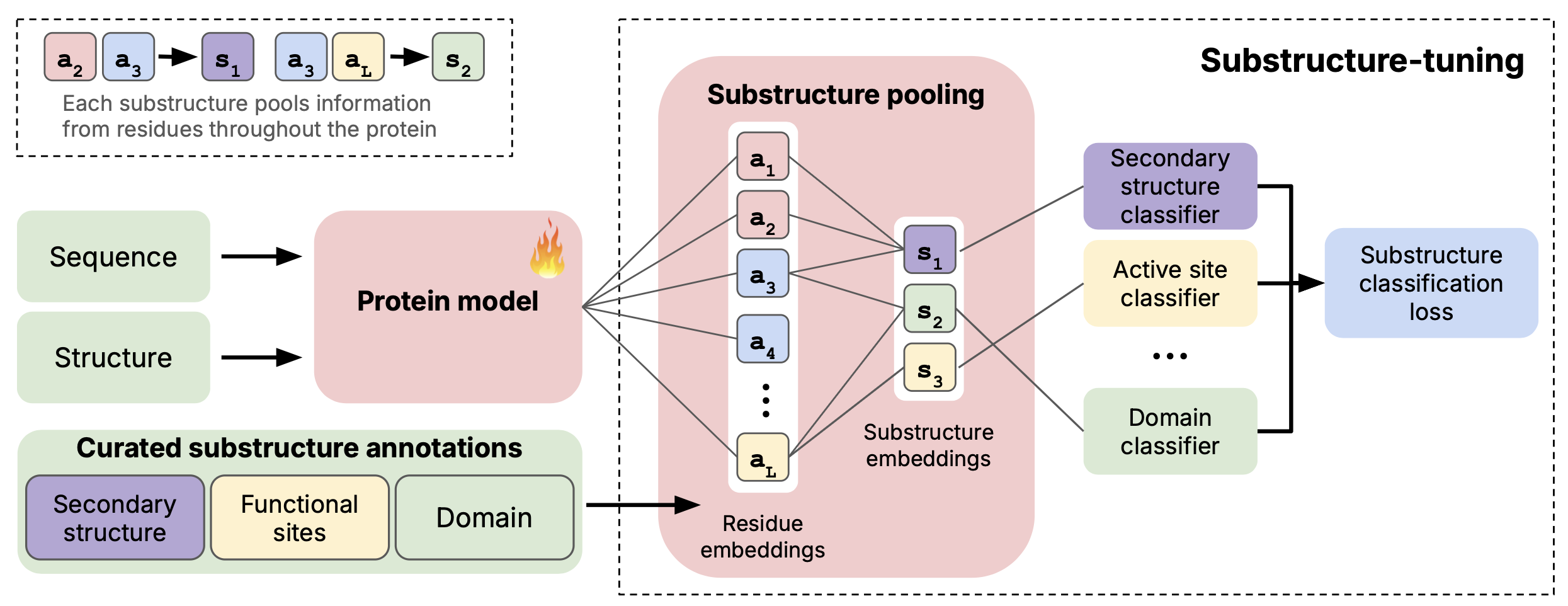
We vary the substructures used for tuning, exploring configurations ranging from small, highly local elements (e.g., active sites spanning < 10 residues) to larger domains, as well as joint training over multiple scales. Substructure-tuning is evaluated on 13 benchmarking tasks using 6 state-of-the-art base models, including both sequence-only and sequence–structure encoders.
Substructure-tuned representations yield consistent improvements on function-related prediction tasks, while effects on localization and residue-level tasks are neutral or negative. Improvements persist even when base models already incorporate global structural inputs, underscoring that substructural signals are distinct and complementary to global protein structure.
Publication
Greater than the Sum of Its Parts: Building Substructure into Protein Encoding Models
Robert Calef, Arthur Liang, Manolis Kellis, Marinka Zitnik
In Review 2025 [arXiv]
@article{calef25greater,
title={Greater than the Sum of Its Parts: Building Substructure into Protein Encoding Models},
author={Calef, Robert and Liang, Arthur and Kellis, Manolis and Zitnik, Marinka},
journal={In Review},
url={https://arxiv.org/abs/2512.18114},
year={2025}
}
Code and Data Availability
Pytorch implementation of Magneton is available in the GitHub repository.