Biomedical reasoning integrates structured, codified knowledge with tacit, experience-driven insights. Depending on the context, quantity, and nature of available evidence, researchers and clinicians use diverse strategies, including rule-based, prototype-based, and case-based reasoning. Effective medical AI models must handle this complexity while ensuring reliability and adaptability.
We introduce KGARevion, a knowledge graph-based agent that answers knowledge-intensive questions. Upon receiving a query, KGARevion generates relevant triplets by leveraging the latent knowledge embedded in a large language model. It then verifies these triplets against a grounded knowledge graph, filtering out errors and retaining only accurate, contextually relevant information for the final answer. This multi-step process strengthens reasoning, adapts to different models of medical inference, and outperforms retrieval-augmented generation-based approaches that lack effective verification mechanisms.
Evaluations on medical QA benchmarks show that KGARevion improves accuracy by over 5.2% over 15 models in handling complex medical queries. To further assess its effectiveness, we curated three new medical QA datasets with varying levels of semantic complexity, where KGARevion improved accuracy by 10.4%. The agent integrates with different LLMs and biomedical knowledge graphs for broad applicability across knowledge-intensive tasks. We evaluated KGARevion on AfriMed-QA, a newly introduced dataset focused on African healthcare, demonstrating its strong zero-shot generalization to underrepresented medical contexts.
Motivation
Medical reasoning involves making diagnostic and therapeutic decisions while also understanding the pathology of diseases. Unlike many other scientific domains, medical reasoning often relies on vertical reasoning, using analogy more heavily. For instance, in biomedical research, an organism such as Drosophila is used as an exemplar to model a disease mechanism, which is then applied by analogy to other organisms, including humans. In clinical practice, the patient serves as an exemplar, with generalizations drawn from many overlapping disease models and similar patient populations. In contrast, fields like physics and chemistry tend to be horizontally organized, where general principles are applied to specific cases. This distinction highlights the unique challenges that medical reasoning poses for question-answering (QA) models.
While large language models (LLMs) have demonstrated strong general capabilities, their responses to medical questions often suffer from incorrect retrieval, missing key information, and misalignment with current scientific and medical knowledge. Additionally, they can struggle to provide contextually relevant answers that account for specific local contexts, such as patient demographics or geography, as well as specific areas of biology. A major issue lies in these models’ inability to systematically integrate different types of evidence. Specifically, they have difficulty combining scientific factual (structured, codified) knowledge derived from formal, rigorous research with tacit (noncodified) knowledge—expertise and lessons learned—which is crucial for contextualizing and interpreting scientific evidence in relation to the specific modifying factors of a given medical question.
LLM-powered QA models often lack such multi-source and grounded knowledge necessary for medical reasoning, which requires understanding the nuanced and specialized nature of medical concepts. Additionally, LLMs trained on general knowledge may struggle to solve medical problems that demand specialized in-domain knowledge. This shortcoming arises from their inability to discern subtle, granular differences that are critical in medical contexts. As a result, LLMs face challenges in complex medical reasoning because such reasoning requires both: 1) simultaneous consideration of dependencies across multiple medical concepts within an input question, and 2) precise, local in-domain knowledge of semantically similar concepts that can carry different medical meanings, as we show in the figure below.
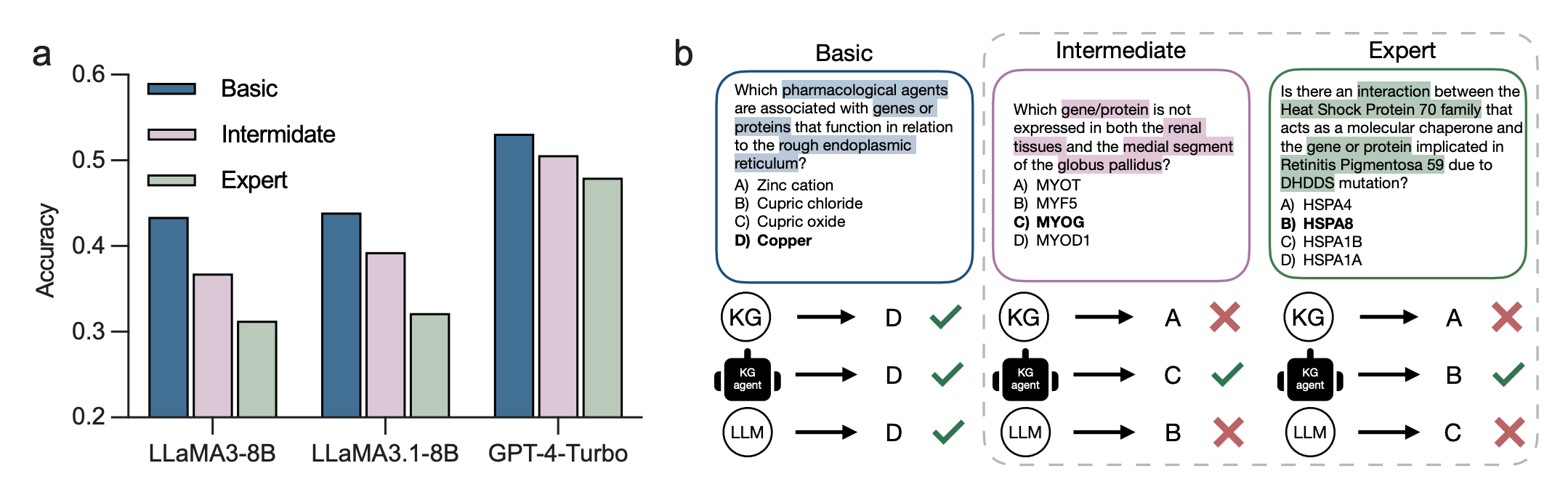
The prevailing strategy to address these challenges is the use of information retrieval techniques, such as retrieval-augmented generation (RAG), which follows a Retrieve-then-Answer paradigm. Although these methods can provide multi-source knowledge from external databases, the accuracy of the generated answers depends heavily on the quality of the retrieved information, making them vulnerable to potential errors. Data repositories and knowledge bases these models draw from contain incomplete or incorrect information, leading to inaccurate retrieval. Further, many RAG-based methods lack post-retrieval verification mechanisms to validate that retrieved information is factually correct and does not miss key information. Knowledge graphs (KGs) of medical concepts have been widely adopted as a grounded knowledge base to provide precise and specialized in-domain knowledge for medical QA models. While KGs can enhance the performance of these models, they are often incomplete. Consequently, approaches that retrieve medical concepts from a KG based solely on the presence of direct associations (edges) between concepts are insufficient. For instance, concepts representing two proteins with distinct biological roles may not be directly connected in the KG, even though these proteins share similar biological representations.
To advance LLM-powered models for knowledge-intensive medical QA, it is essential to develop models that can (1) consider complex associations between several medical concepts at the same time, (2) systematically integrate multi-source knowledge, and (3) effectively verify and ground the retrieved information to ensure contextual relevance and accuracy.
KGARevion: KG-Based Agent for Knowledge-Intensive QA in Medicine
We introduce KGARevion, a knowledge graph-based LLM agent designed for complex medical question answering (QA). KGARevion integrates the non-codified knowledge of LLMs with the structured, codified knowledge embedded in medical concept KGs. It operates through four key actions, as shown in the figure below.
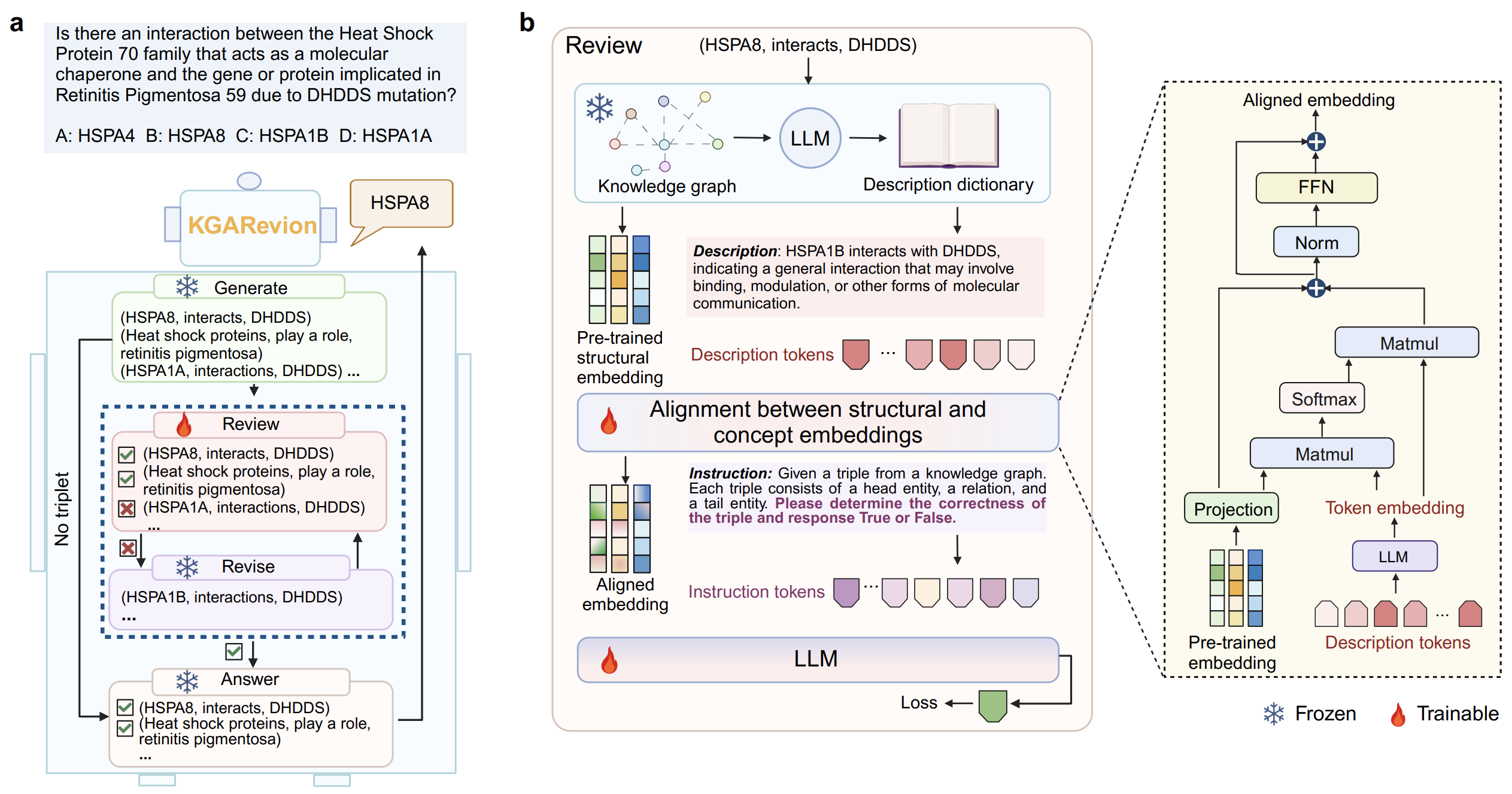
First, KGARevion prompts the LLM to generate relevant triplets based on the input question. To ensure the accuracy of these generated triplets and fully leverage the structured KG, KGARevion fine-tunes the LLM on a KG completion task. This involves incorporating pre-trained structural embeddings of triplets as prefix tokens. The fine-tuned model is then used to evaluate the correctness of the generated triplets.
Next, KGARevion performs a ‘Revise’ action to correct any erroneous triplets, ultimately identifying the correct answer based on the verified triplets. Given the complexity of medical reasoning, KGARevion adaptively selects the most appropriate reasoning strategy for each question, allowing for more nuanced and context-aware QA. This flexibility enables KGARevion to handle both multiple-choice and open-ended questions effectively.
Publication
KGARevion: An AI Agent for Knowledge-Intensive Biomedical QA
Xiaorui Su, Yibo Wang, Shanghua Gao, Xiaolong Liu, Valentina Giunchiglia, Djork-Arné Clevert, Marinka Zitnik
International Conference on Learning Representations, ICLR 2025 [arXiv] [OpenReview]
@article{su2025knowledge,
title={KGARevion: An AI Agent for Knowledge-Intensive Biomedical QA},
author={Su, Xiaorui and Wang, Yibo and Gao, Shanghua and Liu, Xiaolong and Giunchiglia, Valentina and Clevert, Djork-Arn{\'e} and Zitnik, Marinka},
journal={International Conference on Learning Representations, ICLR},
year={2025}
}
Code Availability
Pytorch implementation of KGARevion is available in the GitHub repository.