Spatial perturbation transcriptomics measures how genetic or chemical edits alter gene expression while preserving tissue context. Perturbation outcomes depend on a cell's intrinsic state and also on how effects propagate across cellular microenvironments.
We present CONCERT, a niche-aware generative model that embeds perturbation context and learns spatial kernels with a Gaussian process variational autoencoder to predict perturbation effects across tissue. We formalize three tasks: patch, border, and niche, predicting responses in nearby unperturbed regions, at tissue interfaces, and as a function of surrounding microenvironments. We evaluate CONCERT on Perturb-map lung datasets. CONCERT outperforms state-of-the-art models (dissociated counterfactuals, spatialized perturbation models, and kNN), reducing E-distance by up to 33.77% (patch), 26.05% (border), and 33.74% (niche) versus the next best, with mean absolute error down by up to 23.28% and Pearson correlation up by up to 9.10%.
Two case studies go beyond benchmarking. In dextran sodium sulfate-induced colitis, CONCERT reconstructs spatial gene expression at unmeasured time points, produces longitudinal comparisons across unpaired mice, resolves inter-mouse heterogeneity, and recovers consistent temporal declines of inflammation-associated genes across regions. In ischemic stroke, CONCERT predicts responses under variable lesion sizes and in a 3D formulation across brain sections, capturing lesion-core and peri-lesion patterns. CONCERT performs niche-aware counterfactual prediction, reconstructs missing spatial data, and models perturbation responses across tissues.
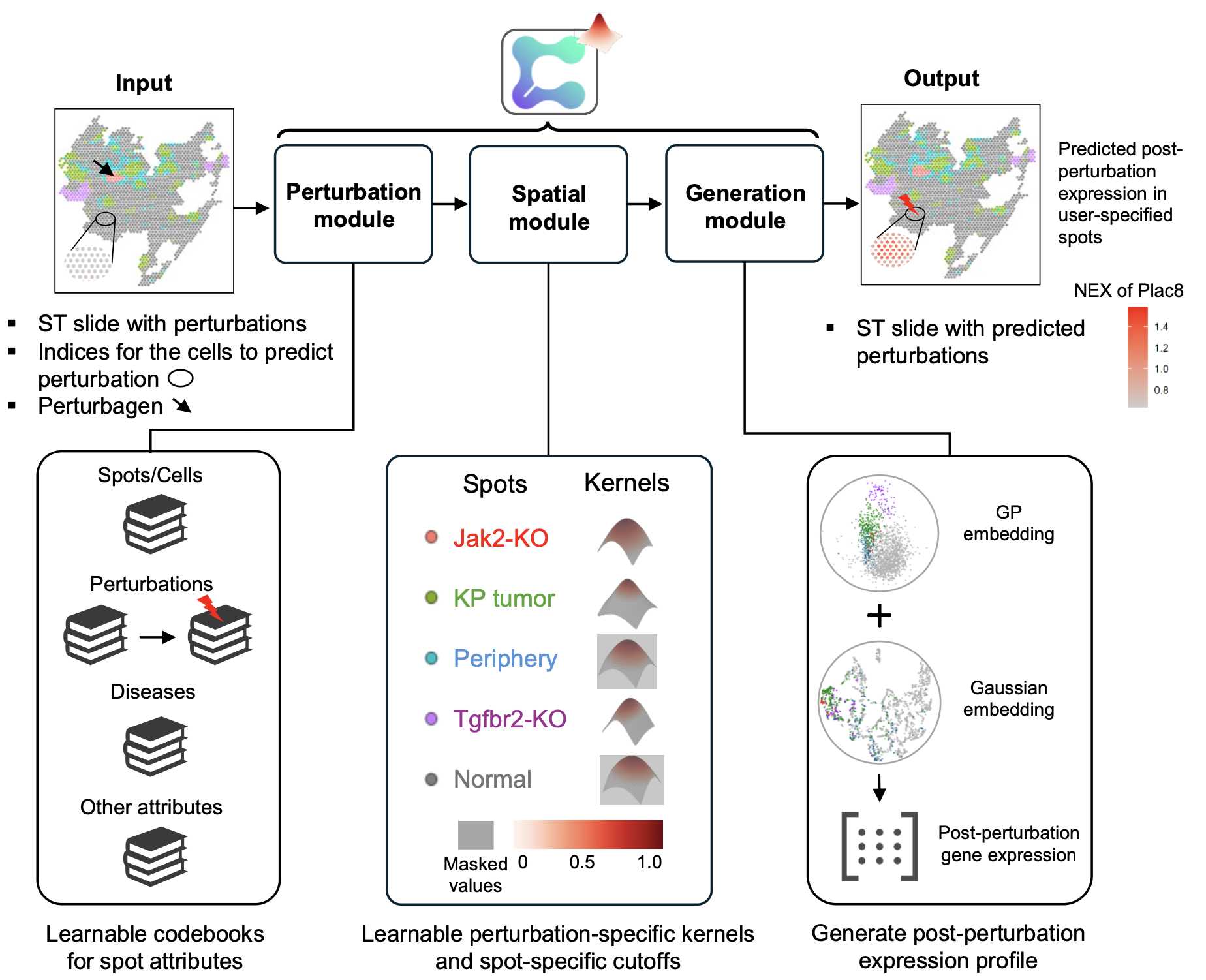
CONCERT: Niche-Aware Virtual Cell Model
CONCERT is a niche-aware generative counterfactual model that predicts how a perturbation’s effect disperses across tissue and alters local gene expression. It takes a tissue’s spatial transcriptomics slide(s) as input—spot/cell gene expression, 2D/3D coordinates, and attributes such as perturbation identity, disease state, dose, lesion size, or time—and outputs predicted post-perturbation expression (rGEX) at user-selected locations, including unseen or imputed spots.
The perturbation module disentangles spot attributes into compact latent codes: categorical variables (e.g., CRISPR knockout, niche, slide ID) are embedded; continuous variables (e.g., time or lesion size) pass through small MLPs, enabling smooth interpolation and “what-if” edits by swapping only the perturbation code while holding cell state and position fixed. The spatial module then propagates the encoded perturbation through tissue with a perturbation-specific Gaussian-process kernel that has learnable, anisotropic length-scales and spot-specific cutoffs. This induces non-local, directionally aware dispersion that respects boundaries and niches rather than copying nearest neighbors. The generation module is a variational decoder that combines latent cell state and the propagated perturbation field to reconstruct rGEX; evaluating it at new coordinates or times enables resolution enhancement, in-painting, temporal interpolation, and 3D propagation across sections.
CONCERT (i) performs counterfactual prediction for patch, border, niche, and cross-niche tasks; (ii) densifies slides and in-paints damaged regions while preserving plausible responses; (iii) interpolates time and lesion size; and (iv) supports 2D and 3D kernels for multi-section tissue. Predictions come with calibrated uncertainty (≈95% coverage in-distribution).
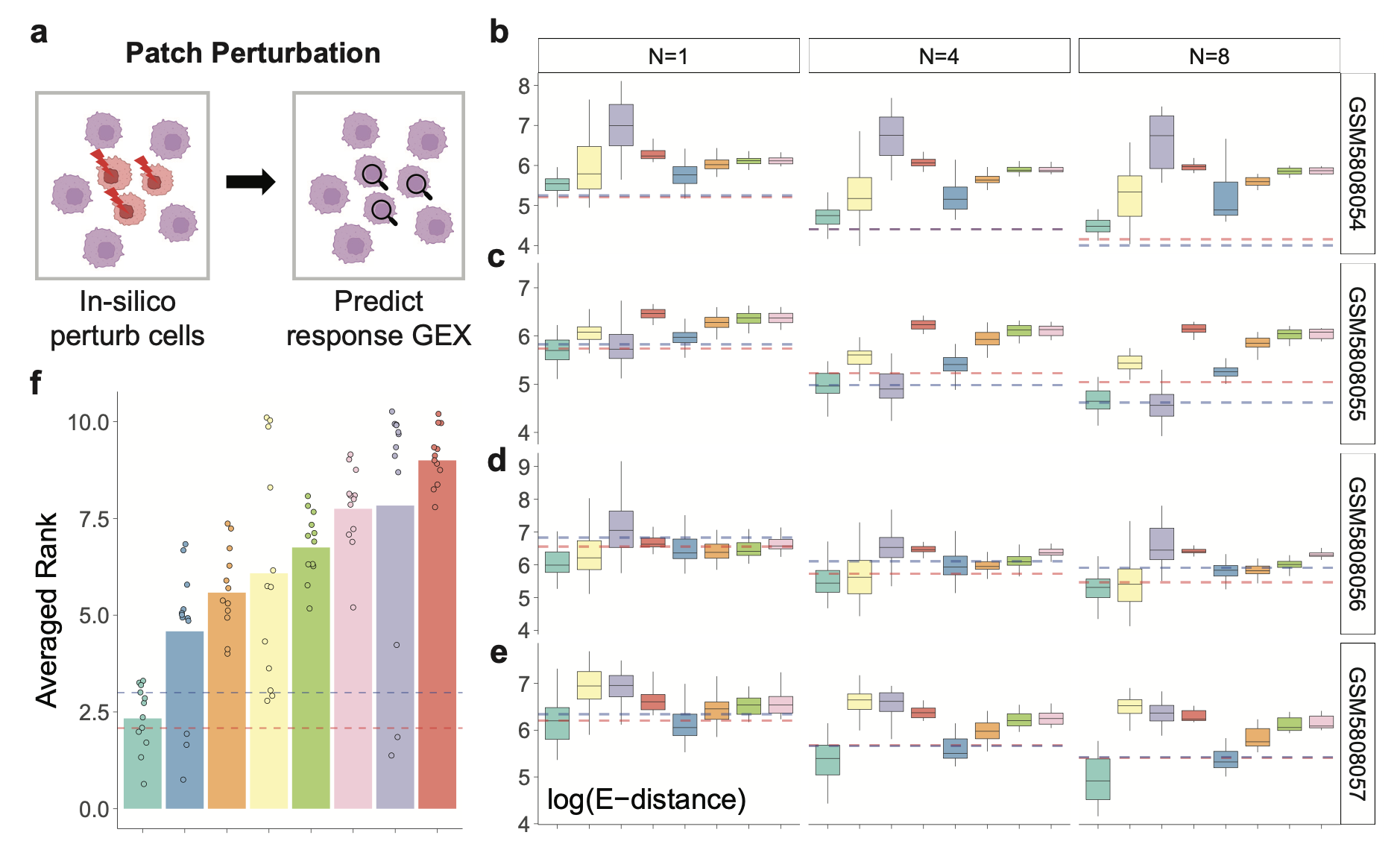
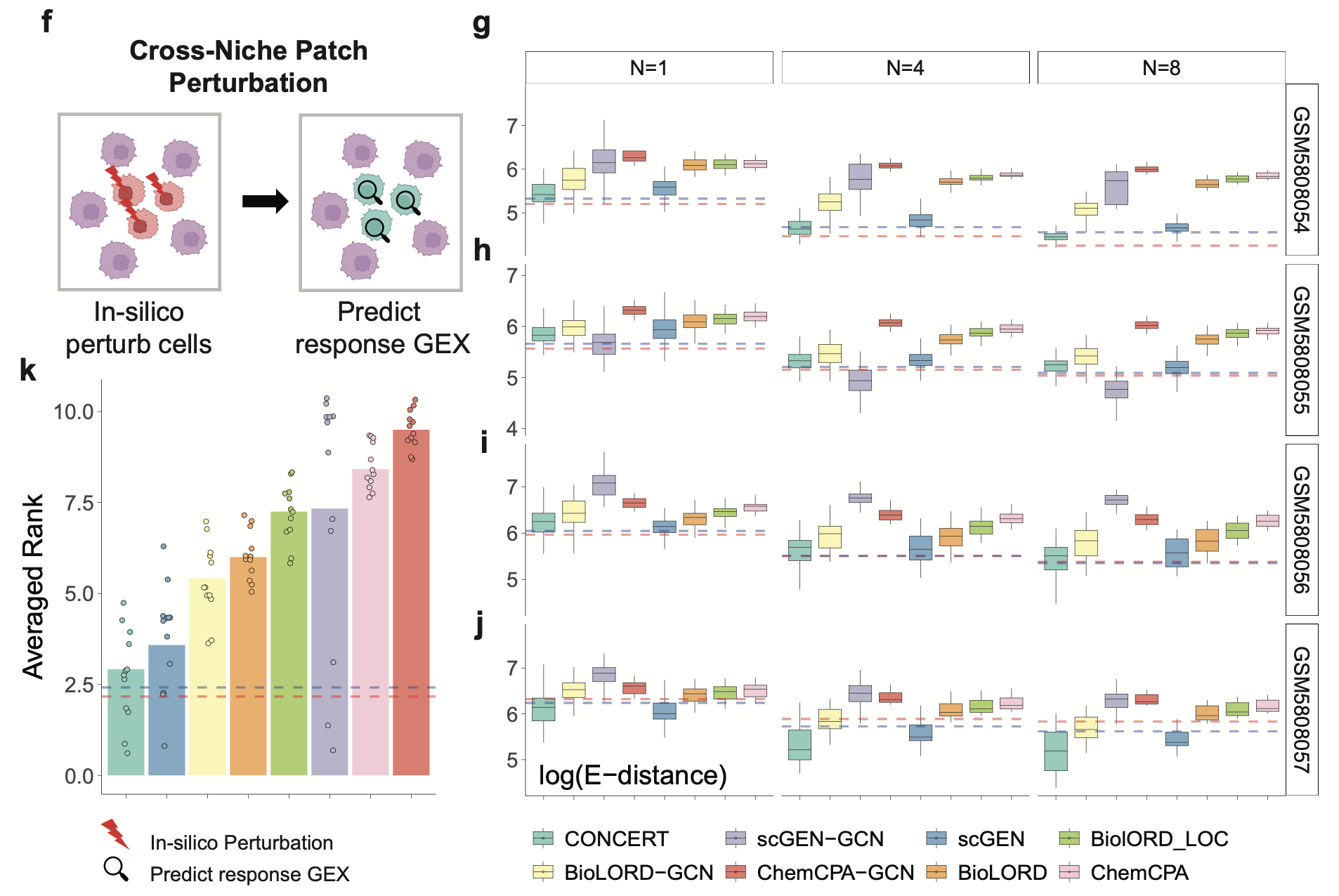
Patch Predictions Within the Same Niche
Given a patch of spots in source state A, predict their rGEX after perturbing them to target state B observed nearby within the same niche. This probes whether a model can reproduce local responses without crossing tissue interfaces.
Across 12 conditions (4 slides × 3 patch sizes), CONCERT achieved the best E-distance in 9/12, closely approaching or surpassing the spatial kNN “upper bound” on multiple slides. Relative to the next-best method, E-distance dropped by up to 33.77%, with consistent gains in MAE and PCC. Average rank was 2.33, near the spatial kNN’s 2.08.
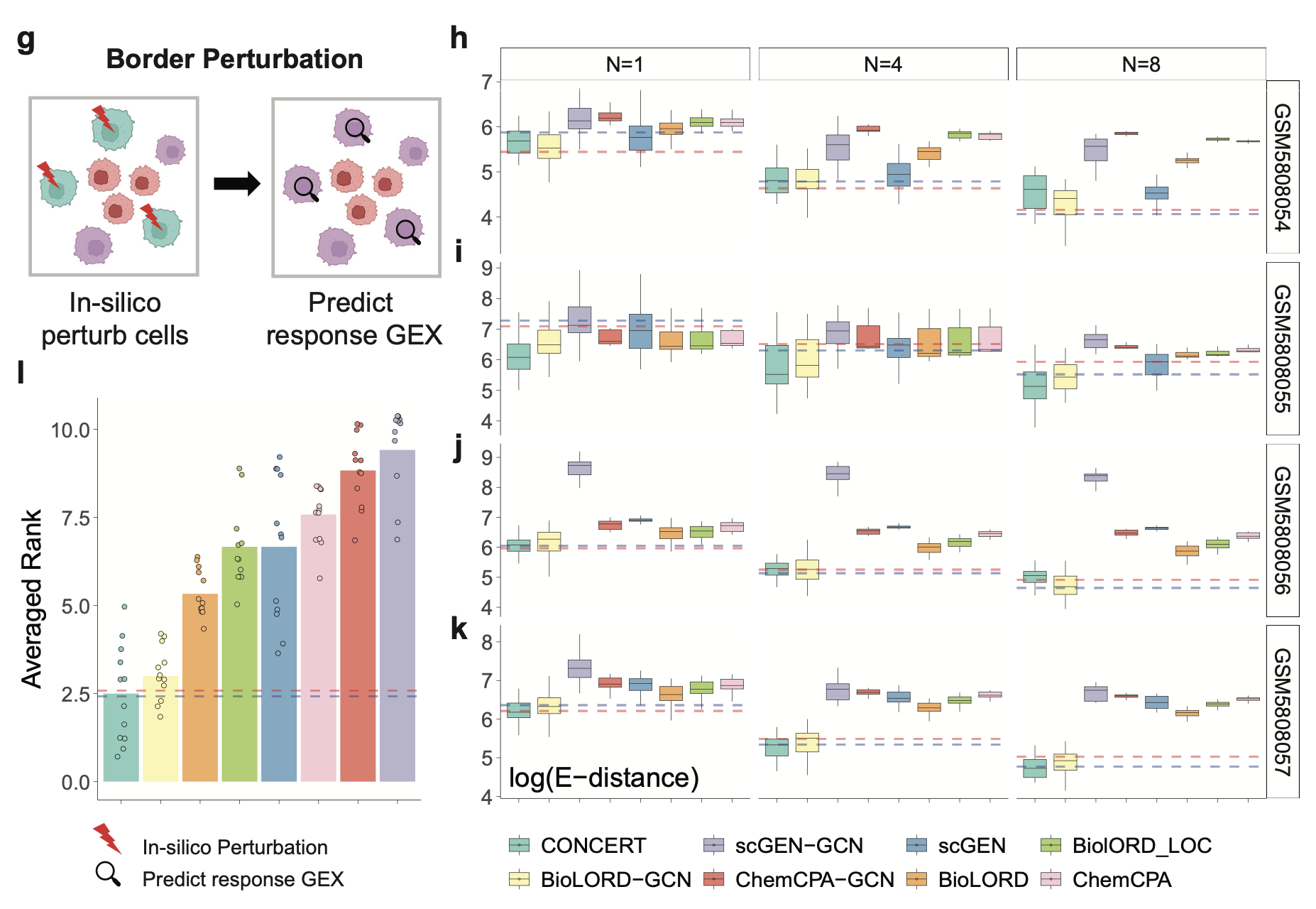
Predictions at Tissue Boundaries
Perturb spots along the edge of a patch and predict their rGEX, a boundary-aware setting with sharp transitions at tissue interfaces.
CONCERT topped 8/12 conditions and reduced E-distance by up to 26.05% versus the next-best model, with an average rank of 2.50, again approaching the spatial kNN bound (2.41). Improvements held across MAE and PCC.
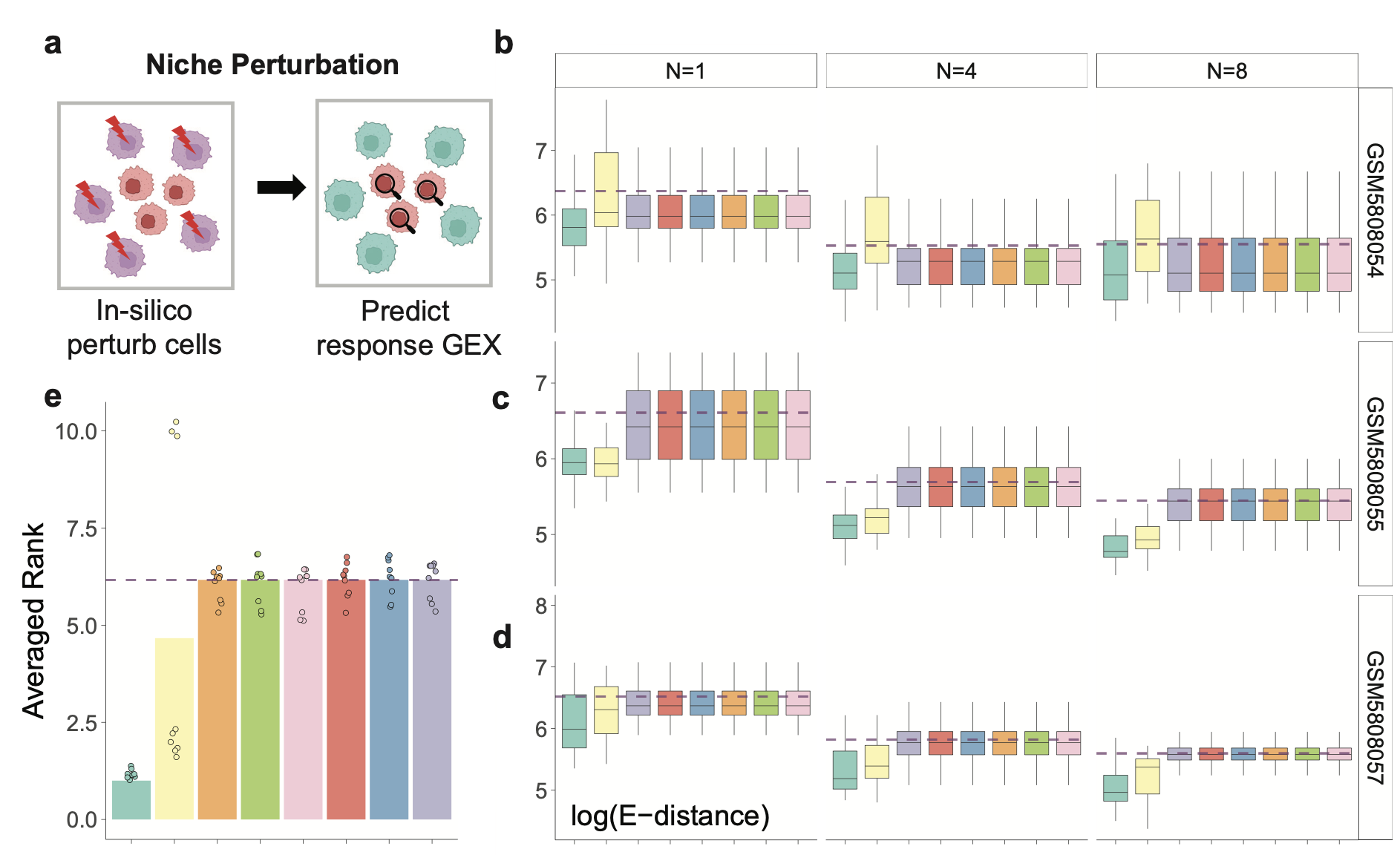
Perturb the Niche, Predict the Core
Perturb the surrounding niche (border) from A to B and predict rGEX inside the core patch—requiring models to capture non-local propagation from neighbors. Only methods that model microenvironmental influence can perform this task.
Only CONCERT and one spatialized baseline could run this task; CONCERT was best in all 9/9 conditions, lowering E-distance by up to 33.74% and achieving the top average rank (1.0) across slides and patch sizes.
Patch Predictions Across Niches
Perturb a patch in state A toward state B that is observed in a different niche, forcing extrapolation across microenvironments.
CONCERT ranked first in 7/12 conditions and reduced E-distance by up to ~24% (with additional significant gains across MAE and PCC), yielding the best overall average rank (2.91) across all tests. Uncertainty calibration was near nominal 95% coverage.
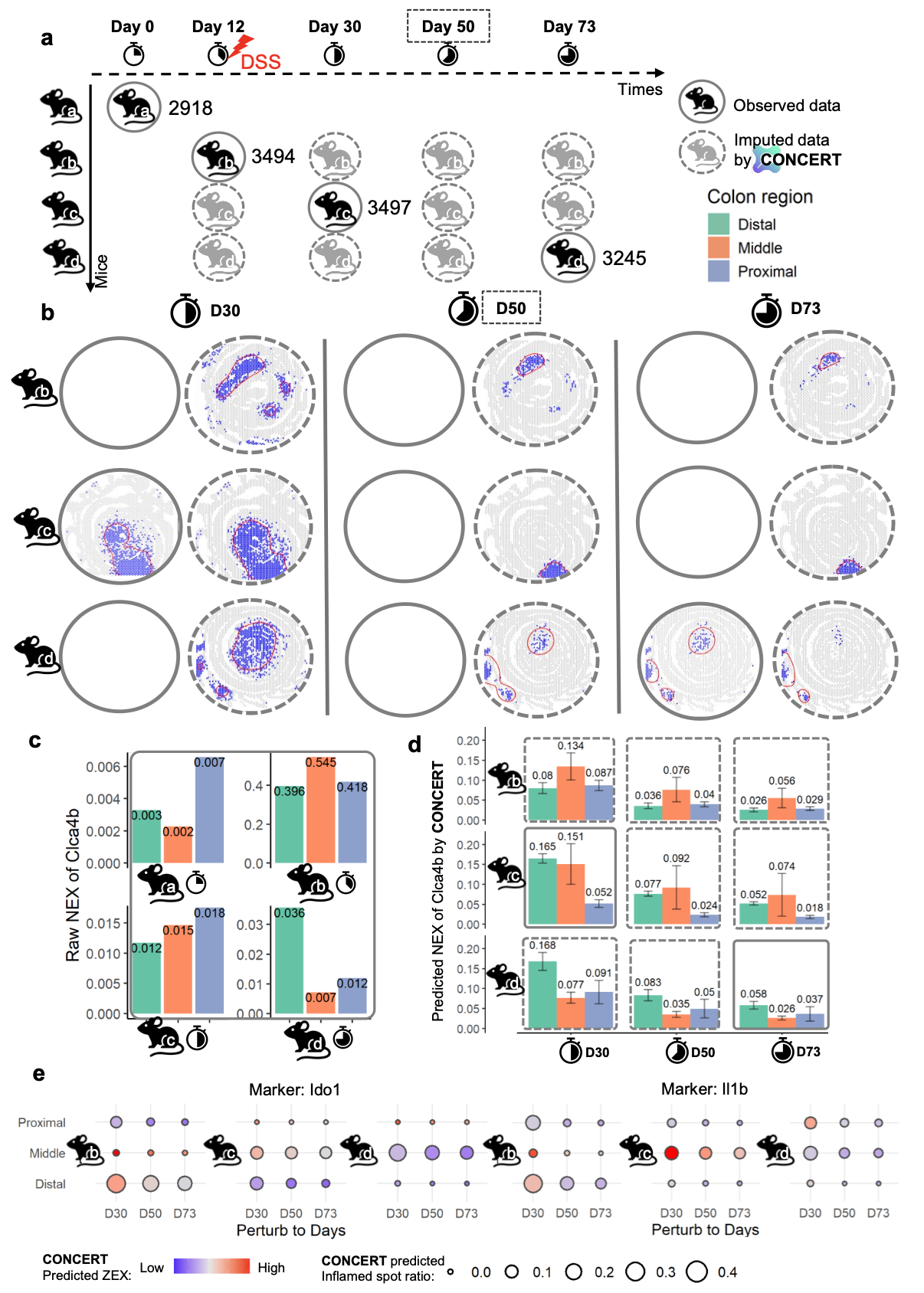
In Vivo Case Studies: Mouse Colitis and Stroke
We asked whether CONCERT can support causal “what-if” reasoning in vivo beyond what experiments can measure or control. Two case studies test distinct axes:
- colitis time course: reconstruct unmeasured time points and enable longitudinal comparisons across different mice; and
- ischemic stroke: simulate location and size-specific lesions in 2D and extend dispersion to 3D across brain sections.
In colitis time course, CONCERT reconstructed spatial rGEX at day 30, an unseen day 50, and day 73 for each mouse, revealing consistent regional declines in inflammation markers (Clca4b, Ido1, Il1b) that were obscured by inter-mouse variability in the raw data. In stroke, CONCERT simulated multi-region ischemia and lesion-size effects on healthy slides, recovering known gene-level patterns (core-maximal Gm42418 and peri-lesion Lcn2/Spp1) and capturing z-axis propagation in a 3D kernel across sections.
These studies demonstrate a practical virtual cell/tissue workflow: fill in missing spatial data, run counterfactual interventions at user-chosen locations, sizes, and times, and compare conditions that are otherwise unpaired or infeasible to acquire. The same engine also supports resolution enhancement and in-painting of damaged tissue while preserving biologically plausible responses.
Publication
CONCERT predicts niche-aware perturbation responses in spatial transcriptomics
Xiang Lin, Zhenglun Kong, Ghosh Soumya, Manolis Kellis, Marinka Zitnik
In Review 2025 [bioRxiv]
@article{lin2025concert,
title={CONCERT predicts niche-aware perturbation responses in spatial transcriptomics},
author={Lin, Xiang and Kong, Zhenglun and Ghosh, Soumya and Kellis, Manolis and Zitnik, Marinka},
journal={In Review},
url={https://www.biorxiv.org/content/10.1101/2025.11.08.686890},
year={2025}
}
Code and Data Availability
Pytorch implementation of CONCERT is available in the GitHub repository.