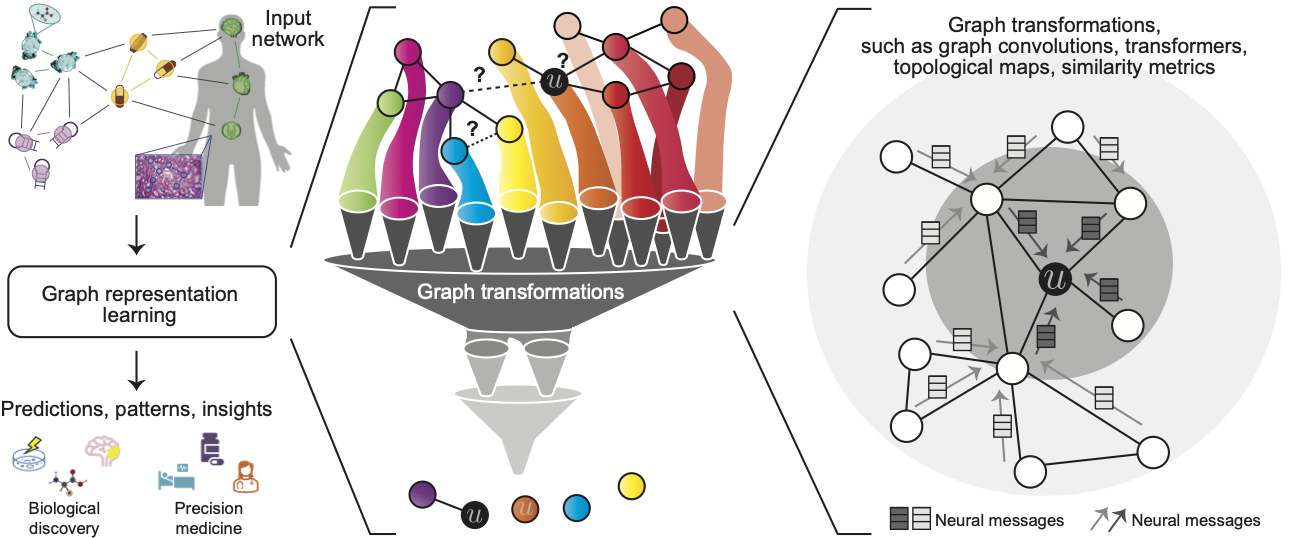
Motivation
Networks (or graphs) are pervasive in biology and medicine, from molecular interaction maps to population-scale social and health interactions. For instance, molecular structure can be translated from atoms and bonds into nodes and edges, respectively; protein interactions naturally form a network based on the existence of a physical interaction or functional relationship; networks can be composed of drugs (e.g., small compounds), proteins, and diseases to allow the modeling of drug-drug interactions, binding of drugs to target proteins, and identification of drug-disease therapeutic opportunities; and patient records can be represented as networks, where edges may indicate co-occurrences of medical codes in health records.
Graph representation learning, also known as machine learning on graphs, geometric deep learning, or graph neural networks (GNNs), has emerged as a leading paradigm for deep learning on networked datasets. Deep learning on graphs is particularly challenging because graphs contain complex topographical structures, no fixed node ordering, and no reference points. Graphs can also comprise many different kinds of entities (nodes) and rich interactions (edges) relating them to each other. Classic deep learning methods cannot consider such diverse structural properties and rich interactions, which are the essence of networks, because they are designed for fixed-size grids (i.e., images and tabular datasets) or sequences (i.e., text). Akin to how deep learning on images and text has revolutionized the image analysis and natural language processing fields, advances in graph representation learning have enabled the scientific community to use deep learning much more broadly, not only for images and text datasets but for any interconnected, networked data system. These algorithmic advances have created new frontiers for applications of deep learning in biology and medicine. Moreover, they have facilitated scientific innovation in biology and medicine, which we will cover in the tutorial. Thus, to further stimulate algorithmic and scientific innovation, our tutorial aims to provide a synthesis and review of graph representation learning in biomedicine that would be accessible to a broad scientific audience.
Program and materials
- (30 min) Part 1: Overview: Introduction to graph representation learning for biomedicine [PDF Materials]
- (60 min) Part 2: Methods: Neural message passing, graph neural networks, equivariant neural networks [PDF Materials]
- (15 min) Break
- (90 min) Part 3: Applications: Precision medicine [PDF Materials]
- Graph representation learning for disease understanding, including methods that inject transcriptomic data into protein interaction networks to identify candidate biomarkers for disease progression, model the effects of non-coding regions on disease, and incorporate non-coding RNA interactions into protein interaction networks.
- Single-cell transcriptomics analysis
- Spatial transcriptomics analysis
- Graph representation learning for therapeutic development, including methods for modeling molecular graphs for small compounds, quantifying drug-drug and drug-target interactions.
- Molecular property prediction, drug-target interaction prediction, molecular generation
- Drug design
- Drug repurposing
- Graph representation learning for patient analyses, focusing specifically on personalizing medical knowledge networks with patient records.
- Histopathology images of tissue biopsies
- Patient electronic health records
- Graph representation learning for disease understanding, including methods that inject transcriptomic data into protein interaction networks to identify candidate biomarkers for disease progression, model the effects of non-coding regions on disease, and incorporate non-coding RNA interactions into protein interaction networks.
- (15 min) Break
- (30 min) Part 4: Demos, practical advice and resources, and hands-on exercises. We will cover the following materials:
- Interactive design of efficacious drugs with deep graph learning [Demo] [DB00503.sdf] [O75469-Nuclear-receptor-subfamily-1-group-I-member-2] [P07550-Beta-2-adrenergic-receptor]
- Practical advice and resources [PDF Materials]
- Applications in therapeutic science using Therapeutic Data Commons, specifically tutorials U1.1 and U1.2 [Tutorials]
Tutorial info
The tutorial was held at the ISMB 2022 conference, July 10-14, 2022, as tutorial VT4 on Thursday, July 7, 9:00 am - 1:00 pm CDT.
The target audiences are graduate students, researchers, scientists and practitioners in both academia and industry who are interested in applying graph machine learning to precision medicine problems.
Tutorial recordings
Presenters
Michelle M. Li is a Ph.D candidate in Biomedical Informatics at Harvard University. Her research focuses on developing deep graph representation learning algorithms that inject specific biological context to enrich predictions in low sample settings (e.g., diagnosing patients with rare diseases, predicting patients’ response to novel drugs). Prior to Harvard, she studied mathematics and computer science at Stanford University, where she also developed bioinformatics and machine learning methods to disentangle the mechanisms of antimicrobial resistance and susceptibility. Her work has been selected for spotlight presentations (IJCAI 2021, ICML 2021) and a best poster award (ICML 2021), and her recently released review paper on graph representation learning for biomedicine—the topic of this tutorial—has been very well received by both the machine learning and computational biology communities.
Marinka Zitnik is an Assistant Professor at Harvard University with appointments in the Department of Biomedical Informatics, Broad Institute of MIT and Harvard, and Harvard Data Science. Dr. Zitnik has published extensively in top ML venues (e.g., NeurIPS, ICLR, ICML) and leading journals (e.g., Nature Methods, Nature Communications, PNAS). This research won best paper and research awards from the International Society for Computational Biology, Bayer Early Excellence in Science Award, Amazon Faculty Research Award, Roche Alliance with Distinguished Scientists Award, Rising Star Award in Electrical Engineering and Computer Science, and Next Generation in Biomedicine Recognition, being the only young scientist who received such recognition in both EECS and Biomedicine.
Other materials
Tutorials on graph machine learning for precision medicine
Tutorials on graph machine learning for therapeutic science
Associated publication
Graph representation learning in biomedicine and healthcare
@article{li2022Graph,
title={Graph Representation Learning in Biomedicine and Healthcare},
author={Li, Michelle M and Huang, Kexin and Zitnik, Marinka},
journal={Nature Biomedical Engineering},
year={2022}
}